Species_specific_genes
Lauren Blake
January 17, 2018
The goal of this script is to explore the categories of genes enriched in the different groups from Cormotif.
Load in data
library("ggplot2")
library("qvalue")
library("RColorBrewer")
library("topGO")Loading required package: BiocGenericsLoading required package: parallel
Attaching package: 'BiocGenerics'The following objects are masked from 'package:parallel':
clusterApply, clusterApplyLB, clusterCall, clusterEvalQ,
clusterExport, clusterMap, parApply, parCapply, parLapply,
parLapplyLB, parRapply, parSapply, parSapplyLBThe following objects are masked from 'package:stats':
IQR, mad, sd, var, xtabsThe following objects are masked from 'package:base':
anyDuplicated, append, as.data.frame, cbind, colMeans,
colnames, colSums, do.call, duplicated, eval, evalq, Filter,
Find, get, grep, grepl, intersect, is.unsorted, lapply,
lengths, Map, mapply, match, mget, order, paste, pmax,
pmax.int, pmin, pmin.int, Position, rank, rbind, Reduce,
rowMeans, rownames, rowSums, sapply, setdiff, sort, table,
tapply, union, unique, unsplit, which, which.max, which.minLoading required package: graphLoading required package: BiobaseWelcome to Bioconductor
Vignettes contain introductory material; view with
'browseVignettes()'. To cite Bioconductor, see
'citation("Biobase")', and for packages 'citation("pkgname")'.Loading required package: GO.dbLoading required package: AnnotationDbiLoading required package: stats4Loading required package: IRangesLoading required package: S4Vectors
Attaching package: 'S4Vectors'The following object is masked from 'package:base':
expand.gridLoading required package: SparseM
Attaching package: 'SparseM'The following object is masked from 'package:base':
backsolve
groupGOTerms: GOBPTerm, GOMFTerm, GOCCTerm environments built.
Attaching package: 'topGO'The following object is masked from 'package:IRanges':
members#library("biomaRt")
library("clusterProfiler")Loading required package: DOSEDOSE v3.4.0 For help: https://guangchuangyu.github.io/DOSE
If you use DOSE in published research, please cite:
Guangchuang Yu, Li-Gen Wang, Guang-Rong Yan, Qing-Yu He. DOSE: an R/Bioconductor package for Disease Ontology Semantic and Enrichment analysis. Bioinformatics 2015, 31(4):608-609clusterProfiler v3.6.0 For help: https://guangchuangyu.github.io/clusterProfiler
If you use clusterProfiler in published research, please cite:
Guangchuang Yu., Li-Gen Wang, Yanyan Han, Qing-Yu He. clusterProfiler: an R package for comparing biological themes among gene clusters. OMICS: A Journal of Integrative Biology. 2012, 16(5):284-287.library("org.Hs.eg.db")library(tidyverse)── Attaching packages ────────────────────────────────── tidyverse 1.2.1 ──✔ tibble 1.4.2 ✔ purrr 0.2.4
✔ tidyr 0.7.2 ✔ dplyr 0.5.0
✔ readr 1.1.1 ✔ stringr 1.3.0
✔ tibble 1.4.2 ✔ forcats 0.2.0── Conflicts ───────────────────────────────────── tidyverse_conflicts() ──
✖ stringr::boundary() masks graph::boundary()
✖ dplyr::collapse() masks IRanges::collapse()
✖ dplyr::combine() masks Biobase::combine(), BiocGenerics::combine()
✖ dplyr::desc() masks IRanges::desc()
✖ tidyr::expand() masks S4Vectors::expand()
✖ dplyr::filter() masks stats::filter()
✖ dplyr::first() masks S4Vectors::first()
✖ dplyr::lag() masks stats::lag()
✖ BiocGenerics::Position() masks ggplot2::Position(), base::Position()
✖ purrr::reduce() masks IRanges::reduce()
✖ dplyr::regroup() masks IRanges::regroup()
✖ dplyr::rename() masks S4Vectors::rename()
✖ dplyr::select() masks AnnotationDbi::select()
✖ purrr::simplify() masks clusterProfiler::simplify()
✖ dplyr::slice() masks IRanges::slice()library(data.table)-------------------------------------------------------------------------data.table + dplyr code now lives in dtplyr.
Please library(dtplyr)!-------------------------------------------------------------------------
Attaching package: 'data.table'The following objects are masked from 'package:dplyr':
between, first, lastThe following object is masked from 'package:purrr':
transposeThe following object is masked from 'package:IRanges':
shiftThe following objects are masked from 'package:S4Vectors':
first, secondlibrary(plyr)-------------------------------------------------------------------------You have loaded plyr after dplyr - this is likely to cause problems.
If you need functions from both plyr and dplyr, please load plyr first, then dplyr:
library(plyr); library(dplyr)-------------------------------------------------------------------------
Attaching package: 'plyr'The following objects are masked from 'package:dplyr':
arrange, count, desc, failwith, id, mutate, rename, summarise,
summarizeThe following object is masked from 'package:purrr':
compactThe following object is masked from 'package:IRanges':
descThe following object is masked from 'package:S4Vectors':
renameThe following object is masked from 'package:graph':
joinlibrary("dplyr")
# Load colors
pal <- c(brewer.pal(9, "Set1"), brewer.pal(8, "Set2"), brewer.pal(12, "Set3"))
# Functions for plots
bjpm<-
theme(
panel.border = element_rect(colour = "black", fill = NA, size = 2),
plot.title = element_text(size = 16, face = "bold"),
axis.text.y = element_text(size = 14,face = "bold",color = "black"),
axis.text.x = element_text(size = 14,face = "bold",color = "black"),
axis.title.y = element_text(size = 14,face = "bold"),
axis.title.x=element_blank(),
legend.text = element_text(size = 14,face = "bold"),
legend.title = element_text(size = 14,face = "bold"),
strip.text.x = element_text(size = 14,face = "bold"),
strip.text.y = element_text(size = 14,face = "bold"),
strip.background = element_rect(colour = "black", size = 2))
bjp<-
theme(
panel.border = element_rect(colour = "black", fill = NA, size = 2),
plot.title = element_text(size = 16, face = "bold"),
axis.text.y = element_text(size = 14,face = "bold",color = "black"),
axis.text.x = element_text(size = 14,face = "bold",color = "black"),
axis.title.y = element_text(size = 14,face = "bold"),
axis.title.x = element_text(size = 14,face = "bold"),
legend.text = element_text(size = 14,face = "bold"),
legend.title = element_text(size = 14,face = "bold"),
strip.text.x = element_text(size = 14,face = "bold"),
strip.text.y = element_text(size = 14,face = "bold"),
strip.background = element_rect(colour = "black", size = 2))
# Load motif data
Table_Motif <- read.csv("/Users/laurenblake/Desktop/Endoderm_TC/ashlar-trial/data/Table_Motif.csv")
dim(Table_Motif)[1] 8004 2Motif 4
true_false <- Table_Motif[,2] == 4
summary(true_false) Mode FALSE TRUE
logical 7817 187 true_false <- as.numeric(true_false)
# Merge ENSG with true/false
test_gene <- as.vector(true_false)
names(test_gene) <- Table_Motif[,1]
# Run topGO
go_data <- new("topGOdata",
ontology = "BP",
allGenes = test_gene,
geneSel = function(allScore){
return(allScore > 0.01)
},
nodeSize = 5,
annotationFun = annFUN.org,
mapping = "org.Hs.eg.db",
ID = "ensembl")
Building most specific GOs ..... ( 8429 GO terms found. )
Build GO DAG topology .......... ( 12447 GO terms and 28743 relations. )
Annotating nodes ............... ( 7115 genes annotated to the GO terms. )# Perform enrichment test
go_test <- runTest(go_data, algorithm = "weight01", statistic = "fisher")
-- Weight01 Algorithm --
the algorithm is scoring 3152 nontrivial nodes
parameters:
test statistic: fisher
Level 17: 2 nodes to be scored (0 eliminated genes)
Level 16: 9 nodes to be scored (0 eliminated genes)
Level 15: 24 nodes to be scored (11 eliminated genes)
Level 14: 50 nodes to be scored (75 eliminated genes)
Level 13: 80 nodes to be scored (299 eliminated genes)
Level 12: 123 nodes to be scored (743 eliminated genes)
Level 11: 205 nodes to be scored (1759 eliminated genes)
Level 10: 315 nodes to be scored (2556 eliminated genes)
Level 9: 402 nodes to be scored (3771 eliminated genes)
Level 8: 414 nodes to be scored (4738 eliminated genes)
Level 7: 477 nodes to be scored (5510 eliminated genes)
Level 6: 450 nodes to be scored (6198 eliminated genes)
Level 5: 311 nodes to be scored (6563 eliminated genes)
Level 4: 182 nodes to be scored (6767 eliminated genes)
Level 3: 86 nodes to be scored (6905 eliminated genes)
Level 2: 21 nodes to be scored (6966 eliminated genes)
Level 1: 1 nodes to be scored (7035 eliminated genes)go_table <- GenTable(go_data, weightFisher = go_test,
orderBy = "weightFisher", ranksOf = "weightFisher",
topNodes = sum(score(go_test) < .01))
go_table GO.ID Term Annotated
1 GO:0045773 positive regulation of axon extension 21
2 GO:0008643 carbohydrate transport 71
3 GO:0086002 cardiac muscle cell action potential inv... 17
4 GO:0046885 regulation of hormone biosynthetic proce... 8
5 GO:0034122 negative regulation of toll-like recepto... 11
6 GO:0086091 regulation of heart rate by cardiac cond... 13
7 GO:0032720 negative regulation of tumor necrosis fa... 14
8 GO:0061298 retina vasculature development in camera... 11
9 GO:0006704 glucocorticoid biosynthetic process 5
10 GO:0019896 axonal transport of mitochondrion 5
11 GO:0071875 adrenergic receptor signaling pathway 5
12 GO:0086103 G-protein coupled receptor signaling pat... 5
13 GO:0007625 grooming behavior 5
14 GO:0031943 regulation of glucocorticoid metabolic p... 5
15 GO:1901841 regulation of high voltage-gated calcium... 5
16 GO:0032691 negative regulation of interleukin-1 bet... 5
17 GO:0061577 calcium ion transmembrane transport via ... 5
18 GO:0045989 positive regulation of striated muscle c... 5
19 GO:0046661 male sex differentiation 65
20 GO:0060412 ventricular septum morphogenesis 16
21 GO:0046849 bone remodeling 29
22 GO:0007586 digestion 23
23 GO:0042755 eating behavior 6
24 GO:0070933 histone H4 deacetylation 6
25 GO:0001946 lymphangiogenesis 6
26 GO:0007409 axonogenesis 212
27 GO:0071108 protein K48-linked deubiquitination 19
28 GO:0048013 ephrin receptor signaling pathway 56
Significant Expected weightFisher
1 5 0.48 9.2e-05
2 5 1.64 0.00031
3 4 0.39 0.00052
4 3 0.18 0.00062
5 3 0.25 0.00173
6 3 0.30 0.00290
7 3 0.32 0.00363
8 3 0.25 0.00499
9 2 0.12 0.00504
10 2 0.12 0.00504
11 2 0.12 0.00504
12 2 0.12 0.00504
13 2 0.12 0.00504
14 2 0.12 0.00504
15 2 0.12 0.00504
16 2 0.12 0.00504
17 2 0.12 0.00504
18 2 0.12 0.00504
19 3 1.50 0.00507
20 3 0.37 0.00540
21 4 0.67 0.00732
22 3 0.53 0.00740
23 2 0.14 0.00745
24 2 0.14 0.00745
25 2 0.14 0.00745
26 12 4.89 0.00749
27 3 0.44 0.00888
28 5 1.29 0.00907sig.genes <- sigGenes(go_data)
goresults <- sapply(go_table$GO.ID, function(x)
{
genes<-genesInTerm(go_data, x)
genes[[1]][genes[[1]] %in% sig.genes]
})Motif 7
true_false <- Table_Motif[,2] == 7
summary(true_false) Mode FALSE TRUE
logical 7318 686 true_false <- as.numeric(true_false)
# Merge ENSG with true/false
test_gene <- as.vector(true_false)
names(test_gene) <- Table_Motif[,1]
# Run topGO
go_data <- new("topGOdata",
ontology = "BP",
allGenes = test_gene,
geneSel = function(allScore){
return(allScore > 0.01)
},
nodeSize = 5,
annotationFun = annFUN.org,
mapping = "org.Hs.eg.db",
ID = "ensembl")
Building most specific GOs ..... ( 8429 GO terms found. )
Build GO DAG topology .......... ( 12447 GO terms and 28743 relations. )
Annotating nodes ............... ( 7115 genes annotated to the GO terms. )# Perform enrichment test
go_test <- runTest(go_data, algorithm = "weight01", statistic = "fisher")
-- Weight01 Algorithm --
the algorithm is scoring 4828 nontrivial nodes
parameters:
test statistic: fisher
Level 19: 1 nodes to be scored (0 eliminated genes)
Level 18: 2 nodes to be scored (0 eliminated genes)
Level 17: 4 nodes to be scored (8 eliminated genes)
Level 16: 12 nodes to be scored (12 eliminated genes)
Level 15: 30 nodes to be scored (23 eliminated genes)
Level 14: 75 nodes to be scored (103 eliminated genes)
Level 13: 138 nodes to be scored (314 eliminated genes)
Level 12: 220 nodes to be scored (933 eliminated genes)
Level 11: 378 nodes to be scored (2086 eliminated genes)
Level 10: 533 nodes to be scored (2971 eliminated genes)
Level 9: 642 nodes to be scored (4180 eliminated genes)
Level 8: 673 nodes to be scored (5179 eliminated genes)
Level 7: 723 nodes to be scored (5837 eliminated genes)
Level 6: 629 nodes to be scored (6360 eliminated genes)
Level 5: 413 nodes to be scored (6642 eliminated genes)
Level 4: 230 nodes to be scored (6811 eliminated genes)
Level 3: 103 nodes to be scored (6920 eliminated genes)
Level 2: 21 nodes to be scored (6973 eliminated genes)
Level 1: 1 nodes to be scored (7035 eliminated genes)go_table <- GenTable(go_data, weightFisher = go_test,
orderBy = "weightFisher", ranksOf = "weightFisher",
topNodes = sum(score(go_test) < .01))
go_table GO.ID Term Annotated
1 GO:0010469 regulation of receptor activity 122
2 GO:0009611 response to wounding 247
3 GO:0048645 animal organ formation 22
4 GO:0045992 negative regulation of embryonic develop... 13
5 GO:2000107 negative regulation of leukocyte apoptot... 13
6 GO:0006477 protein sulfation 5
7 GO:0048671 negative regulation of collateral sprout... 5
8 GO:0035023 regulation of Rho protein signal transdu... 70
9 GO:0070374 positive regulation of ERK1 and ERK2 cas... 55
10 GO:0008285 negative regulation of cell proliferatio... 278
11 GO:0045766 positive regulation of angiogenesis 48
12 GO:0043116 negative regulation of vascular permeabi... 6
13 GO:0050919 negative chemotaxis 21
14 GO:0050927 positive regulation of positive chemotax... 7
15 GO:2001028 positive regulation of endothelial cell ... 7
16 GO:2000352 negative regulation of endothelial cell ... 12
17 GO:0007411 axon guidance 114
18 GO:0055001 muscle cell development 67
19 GO:0097094 craniofacial suture morphogenesis 8
20 GO:0001541 ovarian follicle development 24
21 GO:0050850 positive regulation of calcium-mediated ... 13
22 GO:0003151 outflow tract morphogenesis 29
23 GO:0043433 negative regulation of DNA binding trans... 68
24 GO:0006898 receptor-mediated endocytosis 114
25 GO:0038084 vascular endothelial growth factor signa... 16
26 GO:0002087 regulation of respiratory gaseous exchan... 5
27 GO:2001214 positive regulation of vasculogenesis 5
28 GO:0060572 morphogenesis of an epithelial bud 5
29 GO:0090025 regulation of monocyte chemotaxis 5
30 GO:1902668 negative regulation of axon guidance 5
31 GO:0071363 cellular response to growth factor stimu... 294
32 GO:0001666 response to hypoxia 174
33 GO:0003148 outflow tract septum morphogenesis 10
34 GO:0051895 negative regulation of focal adhesion as... 10
35 GO:0034121 regulation of toll-like receptor signali... 19
36 GO:0072073 kidney epithelium development 48
37 GO:0014074 response to purine-containing compound 49
38 GO:0034114 regulation of heterotypic cell-cell adhe... 6
39 GO:0010960 magnesium ion homeostasis 6
40 GO:0036353 histone H2A-K119 monoubiquitination 6
41 GO:0009065 glutamine family amino acid catabolic pr... 6
42 GO:0031665 negative regulation of lipopolysaccharid... 6
Significant Expected weightFisher
1 31 10.25 2.3e-08
2 37 20.76 4.3e-05
3 8 1.85 0.00018
4 6 1.09 0.00019
5 5 1.09 0.00023
6 4 0.42 0.00023
7 4 0.42 0.00023
8 19 5.88 0.00025
9 13 4.62 0.00049
10 41 23.37 0.00051
11 12 4.03 0.00051
12 4 0.50 0.00065
13 7 1.77 0.00117
14 4 0.59 0.00141
15 4 0.59 0.00141
16 5 1.01 0.00198
17 22 9.58 0.00200
18 13 5.63 0.00256
19 4 0.67 0.00263
20 7 2.02 0.00278
21 5 1.09 0.00300
22 10 2.44 0.00351
23 11 5.72 0.00407
24 20 9.58 0.00478
25 6 1.34 0.00514
26 3 0.42 0.00519
27 3 0.42 0.00519
28 3 0.42 0.00519
29 3 0.42 0.00519
30 3 0.42 0.00519
31 40 24.71 0.00567
32 19 14.62 0.00649
33 4 0.84 0.00688
34 4 0.84 0.00688
35 5 1.60 0.00702
36 7 4.03 0.00703
37 10 4.12 0.00959
38 3 0.50 0.00974
39 3 0.50 0.00974
40 3 0.50 0.00974
41 3 0.50 0.00974
42 3 0.50 0.00974sig.genes <- sigGenes(go_data)Combine Motifs 4+7
true_false <- Table_Motif[,2] == 7 | Table_Motif[,2] == 4
summary(true_false) Mode FALSE TRUE
logical 7131 873 true_false <- as.numeric(true_false)
# Merge ENSG with true/false
test_gene <- as.vector(true_false)
names(test_gene) <- Table_Motif[,1]
# Run topGO
go_data <- new("topGOdata",
ontology = "BP",
allGenes = test_gene,
geneSel = function(allScore){
return(allScore > 0.01)
},
nodeSize = 5,
annotationFun = annFUN.org,
mapping = "org.Hs.eg.db",
ID = "ensembl")
Building most specific GOs ..... ( 8429 GO terms found. )
Build GO DAG topology .......... ( 12447 GO terms and 28743 relations. )
Annotating nodes ............... ( 7115 genes annotated to the GO terms. )# Perform enrichment test
go_test <- runTest(go_data, algorithm = "weight01", statistic = "fisher")
-- Weight01 Algorithm --
the algorithm is scoring 5182 nontrivial nodes
parameters:
test statistic: fisher
Level 19: 1 nodes to be scored (0 eliminated genes)
Level 18: 2 nodes to be scored (0 eliminated genes)
Level 17: 6 nodes to be scored (8 eliminated genes)
Level 16: 17 nodes to be scored (12 eliminated genes)
Level 15: 40 nodes to be scored (33 eliminated genes)
Level 14: 91 nodes to be scored (147 eliminated genes)
Level 13: 153 nodes to be scored (424 eliminated genes)
Level 12: 243 nodes to be scored (1000 eliminated genes)
Level 11: 418 nodes to be scored (2127 eliminated genes)
Level 10: 587 nodes to be scored (3039 eliminated genes)
Level 9: 684 nodes to be scored (4220 eliminated genes)
Level 8: 720 nodes to be scored (5197 eliminated genes)
Level 7: 764 nodes to be scored (5854 eliminated genes)
Level 6: 662 nodes to be scored (6369 eliminated genes)
Level 5: 430 nodes to be scored (6643 eliminated genes)
Level 4: 237 nodes to be scored (6814 eliminated genes)
Level 3: 105 nodes to be scored (6920 eliminated genes)
Level 2: 21 nodes to be scored (6973 eliminated genes)
Level 1: 1 nodes to be scored (7035 eliminated genes)go_table <- GenTable(go_data, weightFisher = go_test,
orderBy = "weightFisher", ranksOf = "weightFisher",
topNodes = sum(score(go_test) < .01))
go_table GO.ID Term Annotated
1 GO:0010469 regulation of receptor activity 122
2 GO:0045773 positive regulation of axon extension 21
3 GO:0048754 branching morphogenesis of an epithelial... 56
4 GO:0009611 response to wounding 247
5 GO:0045766 positive regulation of angiogenesis 48
6 GO:0008285 negative regulation of cell proliferatio... 278
7 GO:0003151 outflow tract morphogenesis 29
8 GO:0001541 ovarian follicle development 24
9 GO:0048645 animal organ formation 22
10 GO:0045992 negative regulation of embryonic develop... 13
11 GO:2000107 negative regulation of leukocyte apoptot... 13
12 GO:0006477 protein sulfation 5
13 GO:0048671 negative regulation of collateral sprout... 5
14 GO:2000352 negative regulation of endothelial cell ... 12
15 GO:0035023 regulation of Rho protein signal transdu... 70
16 GO:0001570 vasculogenesis 36
17 GO:0048771 tissue remodeling 51
18 GO:0043433 negative regulation of DNA binding trans... 68
19 GO:0070374 positive regulation of ERK1 and ERK2 cas... 55
20 GO:0043116 negative regulation of vascular permeabi... 6
21 GO:0031665 negative regulation of lipopolysaccharid... 6
22 GO:0007411 axon guidance 114
23 GO:0001755 neural crest cell migration 14
24 GO:1903364 positive regulation of cellular protein ... 136
25 GO:0003148 outflow tract septum morphogenesis 10
26 GO:0050806 positive regulation of synaptic transmis... 50
27 GO:0014911 positive regulation of smooth muscle cel... 13
28 GO:0060973 cell migration involved in heart develop... 7
29 GO:0050927 positive regulation of positive chemotax... 7
30 GO:0055119 relaxation of cardiac muscle 7
31 GO:2001028 positive regulation of endothelial cell ... 7
32 GO:0007267 cell-cell signaling 590
33 GO:0010862 positive regulation of pathway-restricte... 11
34 GO:0048286 lung alveolus development 11
35 GO:0002686 negative regulation of leukocyte migrati... 11
36 GO:0042981 regulation of apoptotic process 661
37 GO:0050919 negative chemotaxis 21
38 GO:0030182 neuron differentiation 570
39 GO:0022409 positive regulation of cell-cell adhesio... 77
40 GO:0045765 regulation of angiogenesis 89
41 GO:0072659 protein localization to plasma membrane 129
42 GO:0055001 muscle cell development 67
43 GO:0008217 regulation of blood pressure 50
44 GO:0007186 G-protein coupled receptor signaling pat... 222
45 GO:0071385 cellular response to glucocorticoid stim... 19
46 GO:0048806 genitalia development 15
47 GO:0035988 chondrocyte proliferation 8
48 GO:0097094 craniofacial suture morphogenesis 8
49 GO:0015701 bicarbonate transport 8
50 GO:0007275 multicellular organism development 2075
51 GO:0030198 extracellular matrix organization 127
52 GO:0043588 skin development 91
53 GO:0050850 positive regulation of calcium-mediated ... 13
Significant Expected weightFisher
1 37 13.07 1.9e-08
2 10 2.25 2.2e-05
3 15 6.00 0.00021
4 44 26.45 0.00025
5 15 5.14 0.00029
6 48 29.77 0.00031
7 13 3.11 0.00041
8 9 2.57 0.00052
9 9 2.36 0.00058
10 6 1.39 0.00059
11 6 1.39 0.00059
12 4 0.54 0.00060
13 4 0.54 0.00060
14 6 1.29 0.00077
15 21 7.50 0.00079
16 13 3.86 0.00099
17 13 5.46 0.00120
18 16 7.28 0.00131
19 14 5.89 0.00153
20 4 0.64 0.00164
21 4 0.64 0.00164
22 26 12.21 0.00168
23 6 1.50 0.00208
24 12 14.57 0.00219
25 5 1.07 0.00221
26 10 5.35 0.00316
27 5 1.39 0.00350
28 4 0.75 0.00350
29 4 0.75 0.00350
30 4 0.75 0.00350
31 4 0.75 0.00350
32 86 63.19 0.00367
33 5 1.18 0.00370
34 5 1.18 0.00370
35 5 1.18 0.00370
36 98 70.79 0.00442
37 7 2.25 0.00475
38 103 61.05 0.00531
39 15 8.25 0.00569
40 25 9.53 0.00604
41 24 13.82 0.00610
42 18 7.18 0.00618
43 13 5.35 0.00620
44 42 23.78 0.00627
45 7 2.03 0.00636
46 5 1.61 0.00641
47 4 0.86 0.00642
48 4 0.86 0.00642
49 4 0.86 0.00642
50 307 222.23 0.00657
51 27 13.60 0.00725
52 16 9.75 0.00846
53 5 1.39 0.00859#write.csv(go_table, "/Users/laurenblake/Desktop/go_table_47.csv", quote = FALSE, row.names = FALSE)
sig.genes <- sigGenes(go_data)Motif 2
true_false <- Table_Motif[,2] == 2
summary(true_false) Mode FALSE TRUE
logical 6987 1017 true_false <- as.numeric(true_false)
# Merge ENSG with true/false
test_gene <- as.vector(true_false)
names(test_gene) <- Table_Motif[,1]
# Run topGO
go_data <- new("topGOdata",
ontology = "BP",
allGenes = test_gene,
geneSel = function(allScore){
return(allScore > 0.01)
},
nodeSize = 5,
annotationFun = annFUN.org,
mapping = "org.Hs.eg.db",
ID = "ensembl")
Building most specific GOs ..... ( 8429 GO terms found. )
Build GO DAG topology .......... ( 12447 GO terms and 28743 relations. )
Annotating nodes ............... ( 7115 genes annotated to the GO terms. )# Perform enrichment test
go_test <- runTest(go_data, algorithm = "weight01", statistic = "fisher")
-- Weight01 Algorithm --
the algorithm is scoring 5162 nontrivial nodes
parameters:
test statistic: fisher
Level 19: 1 nodes to be scored (0 eliminated genes)
Level 18: 2 nodes to be scored (0 eliminated genes)
Level 17: 7 nodes to be scored (8 eliminated genes)
Level 16: 14 nodes to be scored (12 eliminated genes)
Level 15: 41 nodes to be scored (36 eliminated genes)
Level 14: 92 nodes to be scored (129 eliminated genes)
Level 13: 151 nodes to be scored (433 eliminated genes)
Level 12: 247 nodes to be scored (1026 eliminated genes)
Level 11: 413 nodes to be scored (2182 eliminated genes)
Level 10: 566 nodes to be scored (3047 eliminated genes)
Level 9: 690 nodes to be scored (4230 eliminated genes)
Level 8: 712 nodes to be scored (5149 eliminated genes)
Level 7: 770 nodes to be scored (5856 eliminated genes)
Level 6: 672 nodes to be scored (6374 eliminated genes)
Level 5: 423 nodes to be scored (6650 eliminated genes)
Level 4: 236 nodes to be scored (6824 eliminated genes)
Level 3: 103 nodes to be scored (6917 eliminated genes)
Level 2: 21 nodes to be scored (6973 eliminated genes)
Level 1: 1 nodes to be scored (7035 eliminated genes)go_table <- GenTable(go_data, weightFisher = go_test,
orderBy = "weightFisher", ranksOf = "weightFisher",
topNodes = sum(score(go_test) < .01))
go_table GO.ID Term Annotated
1 GO:0036092 phosphatidylinositol-3-phosphate biosynt... 22
2 GO:0018108 peptidyl-tyrosine phosphorylation 144
3 GO:0051897 positive regulation of protein kinase B ... 54
4 GO:0038111 interleukin-7-mediated signaling pathway 14
5 GO:0045932 negative regulation of muscle contractio... 5
6 GO:0036492 eiF2alpha phosphorylation in response to... 5
7 GO:0043406 positive regulation of MAP kinase activi... 122
8 GO:0006084 acetyl-CoA metabolic process 20
9 GO:0001708 cell fate specification 20
10 GO:0060326 cell chemotaxis 68
11 GO:0006954 inflammatory response 171
12 GO:0030837 negative regulation of actin filament po... 28
13 GO:0042347 negative regulation of NF-kappaB import ... 11
14 GO:0055081 anion homeostasis 12
15 GO:0120033 negative regulation of plasma membrane b... 12
16 GO:1900027 regulation of ruffle assembly 13
17 GO:0071625 vocalization behavior 6
18 GO:0060315 negative regulation of ryanodine-sensiti... 6
19 GO:0007610 behavior 189
20 GO:0046854 phosphatidylinositol phosphorylation 46
21 GO:0001657 ureteric bud development 35
22 GO:0007034 vacuolar transport 83
23 GO:0021952 central nervous system projection neuron... 10
24 GO:0030168 platelet activation 62
25 GO:0003009 skeletal muscle contraction 14
26 GO:0050777 negative regulation of immune response 41
27 GO:0042593 glucose homeostasis 108
28 GO:0008037 cell recognition 43
29 GO:0019395 fatty acid oxidation 48
30 GO:1901385 regulation of voltage-gated calcium chan... 12
31 GO:0043011 myeloid dendritic cell differentiation 7
32 GO:0022038 corpus callosum development 7
33 GO:0048820 hair follicle maturation 7
34 GO:0051639 actin filament network formation 7
35 GO:1901203 positive regulation of extracellular mat... 7
36 GO:0002250 adaptive immune response 92
37 GO:0017158 regulation of calcium ion-dependent exoc... 29
38 GO:0044827 modulation by host of viral genome repli... 10
39 GO:0033574 response to testosterone 15
40 GO:0007173 epidermal growth factor receptor signali... 70
41 GO:0051171 regulation of nitrogen compound metaboli... 2757
42 GO:0001764 neuron migration 60
43 GO:0014068 positive regulation of phosphatidylinosi... 25
Significant Expected weightFisher
1 12 2.79 3.1e-06
2 27 18.28 0.00039
3 16 6.85 0.00076
4 7 1.78 0.00079
5 4 0.63 0.00116
6 4 0.63 0.00116
7 23 15.48 0.00126
8 6 2.54 0.00131
9 8 2.54 0.00201
10 13 8.63 0.00210
11 29 21.70 0.00250
12 10 3.55 0.00261
13 5 1.40 0.00312
14 5 1.52 0.00312
15 5 1.52 0.00312
16 5 1.65 0.00313
17 4 0.76 0.00313
18 4 0.76 0.00313
19 26 23.99 0.00314
20 13 5.84 0.00370
21 12 4.44 0.00455
22 15 10.53 0.00471
23 5 1.27 0.00473
24 16 7.87 0.00475
25 6 1.78 0.00499
26 9 5.20 0.00500
27 23 13.71 0.00587
28 13 5.46 0.00645
29 10 6.09 0.00655
30 5 1.52 0.00656
31 4 0.89 0.00657
32 4 0.89 0.00657
33 4 0.89 0.00657
34 4 0.89 0.00657
35 4 0.89 0.00657
36 22 11.68 0.00712
37 7 3.68 0.00736
38 4 1.27 0.00737
39 6 1.90 0.00744
40 15 8.88 0.00767
41 326 349.90 0.00843
42 13 7.61 0.00865
43 8 3.17 0.00968#write.csv(go_table, "/Users/laurenblake/Desktop/go_table_2.csv", quote = FALSE, row.names = FALSE)
sig.genes <- sigGenes(go_data)Motif 3+5
true_false <- Table_Motif[,2] == 3 | Table_Motif[,2] == 5
summary(true_false) Mode FALSE TRUE
logical 7165 839 true_false <- as.numeric(true_false)
# Merge ENSG with true/false
test_gene <- as.vector(true_false)
names(test_gene) <- Table_Motif[,1]
# Run topGO
go_data <- new("topGOdata",
ontology = "BP",
allGenes = test_gene,
geneSel = function(allScore){
return(allScore > 0.01)
},
nodeSize = 5,
annotationFun = annFUN.org,
mapping = "org.Hs.eg.db",
ID = "ensembl")
Building most specific GOs ..... ( 8429 GO terms found. )
Build GO DAG topology .......... ( 12447 GO terms and 28743 relations. )
Annotating nodes ............... ( 7115 genes annotated to the GO terms. )# Perform enrichment test
go_test <- runTest(go_data, algorithm = "weight01", statistic = "fisher")
-- Weight01 Algorithm --
the algorithm is scoring 4904 nontrivial nodes
parameters:
test statistic: fisher
Level 19: 1 nodes to be scored (0 eliminated genes)
Level 18: 1 nodes to be scored (0 eliminated genes)
Level 17: 3 nodes to be scored (8 eliminated genes)
Level 16: 13 nodes to be scored (11 eliminated genes)
Level 15: 32 nodes to be scored (23 eliminated genes)
Level 14: 93 nodes to be scored (122 eliminated genes)
Level 13: 154 nodes to be scored (379 eliminated genes)
Level 12: 242 nodes to be scored (1072 eliminated genes)
Level 11: 376 nodes to be scored (2207 eliminated genes)
Level 10: 522 nodes to be scored (3117 eliminated genes)
Level 9: 652 nodes to be scored (4241 eliminated genes)
Level 8: 690 nodes to be scored (5161 eliminated genes)
Level 7: 719 nodes to be scored (5842 eliminated genes)
Level 6: 647 nodes to be scored (6380 eliminated genes)
Level 5: 417 nodes to be scored (6654 eliminated genes)
Level 4: 224 nodes to be scored (6824 eliminated genes)
Level 3: 96 nodes to be scored (6921 eliminated genes)
Level 2: 21 nodes to be scored (6969 eliminated genes)
Level 1: 1 nodes to be scored (7035 eliminated genes)go_table <- GenTable(go_data, weightFisher = go_test,
orderBy = "weightFisher", ranksOf = "weightFisher",
topNodes = sum(score(go_test) < .01))
go_table GO.ID Term Annotated
1 GO:0051301 cell division 366
2 GO:0007062 sister chromatid cohesion 87
3 GO:0032201 telomere maintenance via semi-conservati... 17
4 GO:0034080 CENP-A containing nucleosome assembly 20
5 GO:0006270 DNA replication initiation 31
6 GO:0006266 DNA ligation 15
7 GO:0006297 nucleotide-excision repair, DNA gap fill... 13
8 GO:0071897 DNA biosynthetic process 130
9 GO:0007088 regulation of mitotic nuclear division 99
10 GO:0006273 lagging strand elongation 6
11 GO:0034501 protein localization to kinetochore 9
12 GO:1904874 positive regulation of telomerase RNA lo... 9
13 GO:0045740 positive regulation of DNA replication 32
14 GO:0000083 regulation of transcription involved in ... 21
15 GO:0060236 regulation of mitotic spindle organizati... 25
16 GO:0008283 cell proliferation 824
17 GO:0035404 histone-serine phosphorylation 7
18 GO:0032467 positive regulation of cytokinesis 18
19 GO:0006298 mismatch repair 18
20 GO:0006271 DNA strand elongation involved in DNA re... 11
21 GO:1902969 mitotic DNA replication 8
22 GO:0031126 snoRNA 3'-end processing 5
23 GO:0035405 histone-threonine phosphorylation 5
24 GO:0016925 protein sumoylation 59
25 GO:0007059 chromosome segregation 217
26 GO:0042769 DNA damage response, detection of DNA da... 25
27 GO:0007077 mitotic nuclear envelope disassembly 30
28 GO:0001556 oocyte maturation 12
29 GO:0006977 DNA damage response, signal transduction... 41
30 GO:0000732 strand displacement 21
31 GO:0042276 error-prone translesion synthesis 13
32 GO:0000082 G1/S transition of mitotic cell cycle 153
33 GO:0000076 DNA replication checkpoint 13
34 GO:0044806 G-quadruplex DNA unwinding 6
35 GO:1904851 positive regulation of establishment of ... 6
36 GO:0000731 DNA synthesis involved in DNA repair 49
37 GO:0045132 meiotic chromosome segregation 41
38 GO:0045638 negative regulation of myeloid cell diff... 20
39 GO:0031297 replication fork processing 24
40 GO:0048146 positive regulation of fibroblast prolif... 24
41 GO:0006296 nucleotide-excision repair, DNA incision... 24
42 GO:0032508 DNA duplex unwinding 54
43 GO:0007569 cell aging 58
44 GO:0030574 collagen catabolic process 20
45 GO:0007019 microtubule depolymerization 18
46 GO:0042255 ribosome assembly 39
47 GO:0034475 U4 snRNA 3'-end processing 7
48 GO:0016446 somatic hypermutation of immunoglobulin ... 7
49 GO:0070987 error-free translesion synthesis 11
50 GO:0045143 homologous chromosome segregation 22
51 GO:0000079 regulation of cyclin-dependent protein s... 49
52 GO:0031145 anaphase-promoting complex-dependent cat... 63
53 GO:0007018 microtubule-based movement 142
54 GO:0006260 DNA replication 198
55 GO:0090307 mitotic spindle assembly 42
56 GO:0000212 meiotic spindle organization 8
57 GO:0010826 negative regulation of centrosome duplic... 8
58 GO:0071392 cellular response to estradiol stimulus 8
59 GO:0051988 regulation of attachment of spindle micr... 8
60 GO:0042594 response to starvation 88
61 GO:0042273 ribosomal large subunit biogenesis 42
62 GO:0045648 positive regulation of erythrocyte diffe... 13
63 GO:0048025 negative regulation of mRNA splicing, vi... 18
64 GO:1900182 positive regulation of protein localizat... 57
65 GO:0006283 transcription-coupled nucleotide-excisio... 53
Significant Expected weightFisher
1 91 39.20 5.1e-12
2 30 9.32 2.8e-09
3 12 1.82 7.8e-09
4 12 2.14 1.2e-07
5 15 3.32 1.4e-07
6 9 1.61 6.2e-07
7 8 1.39 1.3e-05
8 39 13.92 1.6e-05
9 30 10.60 4.8e-05
10 5 0.64 7.6e-05
11 6 0.96 9.4e-05
12 6 0.96 9.4e-05
13 14 3.43 0.00015
14 9 2.25 0.00016
15 8 2.68 0.00021
16 115 88.25 0.00023
17 5 0.75 0.00024
18 8 1.93 0.00027
19 8 1.93 0.00027
20 9 1.18 0.00058
21 5 0.86 0.00059
22 4 0.54 0.00060
23 4 0.54 0.00060
24 15 6.32 0.00061
25 69 23.24 0.00066
26 9 2.68 0.00074
27 10 3.21 0.00076
28 6 1.29 0.00077
29 12 4.39 0.00087
30 8 2.25 0.00095
31 6 1.39 0.00131
32 37 16.39 0.00156
33 5 1.39 0.00164
34 4 0.64 0.00164
35 4 0.64 0.00164
36 17 5.25 0.00179
37 15 4.39 0.00212
38 6 2.14 0.00221
39 8 2.57 0.00257
40 8 2.57 0.00257
41 8 2.57 0.00257
42 14 5.78 0.00278
43 11 6.21 0.00315
44 7 2.14 0.00348
45 6 1.93 0.00349
46 8 4.18 0.00349
47 4 0.75 0.00350
48 4 0.75 0.00350
49 5 1.18 0.00370
50 8 2.36 0.00445
51 11 5.25 0.00456
52 14 6.75 0.00585
53 25 15.21 0.00614
54 61 21.21 0.00619
55 12 4.50 0.00620
56 4 0.86 0.00642
57 4 0.86 0.00642
58 4 0.86 0.00642
59 4 0.86 0.00642
60 12 9.42 0.00655
61 9 4.50 0.00854
62 5 1.39 0.00859
63 6 1.93 0.00886
64 10 6.10 0.00887
65 12 5.68 0.00889#write.csv(go_table, "/Users/laurenblake/Desktop/go_table_35.csv", quote = FALSE, row.names = FALSE)
sig.genes <- sigGenes(go_data)Compare enrichments from multiple categories
# Make the different cluster (motif 2, motif 3+5, motif 4+7)
true_false_2 <- Table_Motif[,2] == 2
true_false_35 <- Table_Motif[,2] == 3 | Table_Motif[,2] == 5
true_false_47 <- Table_Motif[,2] == 4 | Table_Motif[,2] == 7
all_df <- data.frame(ensg = Table_Motif[,1], group2 = true_false_2, group35 = true_false_35, group47 = true_false_47)
dim(all_df)[1] 8004 4# Subset to only the ones that don't have 3 FALSE
test_df <- all_df[which(all_df$group2 == "TRUE" | all_df$group35 == "TRUE" | all_df$group47 == "TRUE"), ]
summary(test_df) ensg group2 group35 group47
ENSG00000000460: 1 Mode :logical Mode :logical Mode :logical
ENSG00000001036: 1 FALSE:1712 FALSE:1890 FALSE:1856
ENSG00000001630: 1 TRUE :1017 TRUE :839 TRUE :873
ENSG00000003056: 1
ENSG00000003400: 1
ENSG00000003509: 1
(Other) :2723 # Look at one cluster
formula_res <- compareCluster(ensg~group2, data=test_df, fun="enrichGO", universe = all_df$ensg,
OrgDb = org.Hs.eg.db,
keyType = 'ENSEMBL',
ont = "BP",
pAdjustMethod = "fdr",
qvalueCutoff = 0.05,
maxGSSize = 3000,
minGSSize = 3)
formula_res %>%as.data.frame()%>%select(-geneID)->enrichment.res
row.names(enrichment.res)=NULL
# Compare the clusters
formula_res <- compareCluster(ensg~group2+group35+group47, data=test_df, fun="enrichGO", universe = all_df$ensg,
OrgDb = org.Hs.eg.db,
keyType = 'ENSEMBL',
ont = "BP",
pAdjustMethod = "fdr",
qvalueCutoff = 0.05,
maxGSSize = 3000,
minGSSize = 3)
formula_res %>%as.data.frame()%>%select(-geneID)->enrichment.res
row.names(enrichment.res)=NULL
summary(enrichment.res) Cluster group2 group35
FALSE.FALSE.TRUE:266 Length:464 Length:464
FALSE.TRUE.FALSE:197 Class :character Class :character
TRUE.FALSE.FALSE: 1 Mode :character Mode :character
group47 ID Description
Length:464 Length:464 Length:464
Class :character Class :character Class :character
Mode :character Mode :character Mode :character
GeneRatio BgRatio pvalue
Length:464 Length:464 Min. :0.000e+00
Class :character Class :character 1st Qu.:2.470e-06
Mode :character Mode :character Median :1.580e-04
Mean :4.738e-04
3rd Qu.:7.697e-04
Max. :2.243e-03
p.adjust qvalue Count
Min. :0.0000000 Min. :0.0000000 Min. : 3.00
1st Qu.:0.0002403 1st Qu.:0.0002193 1st Qu.: 13.00
Median :0.0074010 Median :0.0066090 Median : 27.00
Mean :0.0142467 Mean :0.0129641 Mean : 50.08
3rd Qu.:0.0248547 3rd Qu.:0.0227524 3rd Qu.: 61.00
Max. :0.0499949 Max. :0.0467292 Max. :373.00 #Plot
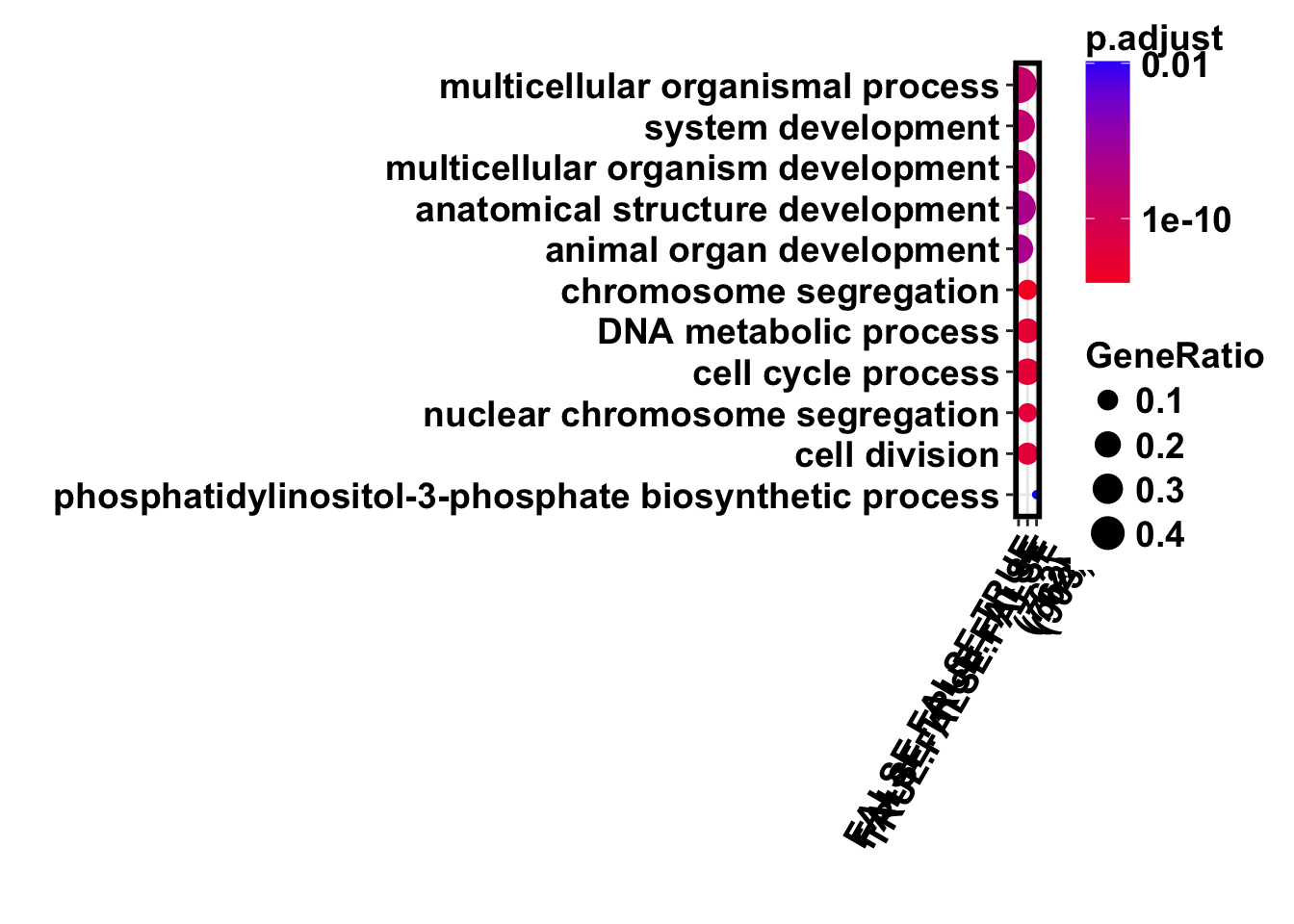
my_breaks = c(0.01,10^-10,10^-20, 10^-30)
dotplot(formula_res)+bjp+theme(axis.text.x = element_text(angle = 60, hjust = 1))+
scale_color_gradient(low ="#f70028",high = "#0200ff",trans="log",breaks = my_breaks, labels = my_breaks)Scale for 'colour' is already present. Adding another scale for
'colour', which will replace the existing scale.