getwd()
## [1] "/Users/laurenblake/Desktop/Regulatory_Evol/ashlar-trial/analysis"
# import sample labels
samples <- read.delim("../../../Reg_Evo_Primates/data/Sample_info_RNAseq_RIN.txt")
RIN <- samples$RIN
# expression
exprs <- read.table("../../../Reg_Evo_Primates/data/250_exp_avg_methyl_hcr_4155_genes.txt", sep="")
# Normalized gene expression data
cpm.voom.cyclic <- readRDS("../../../Reg_Evo_Primates/data/human_chimp_orth_cpm_voom_cyclic.rds")
# Load libraries
library("edgeR")
## Loading required package: limma
library("limma")
library("plyr")
library("ashr")
library("cowplot")
## Warning: package 'cowplot' was built under R version 3.4.4
## Loading required package: ggplot2
## Warning: package 'ggplot2' was built under R version 3.4.4
##
## Attaching package: 'cowplot'
## The following object is masked from 'package:ggplot2':
##
## ggsave
library("vashr")
## Loading required package: SQUAREM
## Loading required package: qvalue
library("devtools")
## Warning: package 'devtools' was built under R version 3.4.4
Set FDR level/svalue
FDR_level <- 0.05
Human heart-kidney
# Check parameters
human_chimp_heart <- c(17, 20, 21, 24, 25, 28, 29)
# Methylation
# Methylation
hc_exprs <- exprs[,2:48]
exprs_methyl <- exprs[,49:95]
# HC heart only
hc_exprs_heart <- hc_exprs[,human_chimp_heart]
methyl <- exprs_methyl[,human_chimp_heart]
rownames(hc_exprs_heart) <- exprs[,1]
rownames(methyl) <- exprs[,1]
Y <- hc_exprs_heart
two_species <- samples$Tissue[human_chimp_heart]
X <- droplevels.factor(two_species)
M <- methyl
RIN_subset <- RIN[human_chimp_heart]
Human heart versus kidney- DE genes
# Run the linear model in limma
design_1 <- model.matrix(~ X + RIN_subset)
fit_all <- lmFit(cpm.voom.cyclic[,human_chimp_heart], design_1)
fit_all <- eBayes(fit_all)
# Get results
HvC_Heart_fit_all = topTable(fit_all, coef=2, adjust="BH", number=Inf, sort.by="none")
HvC_Heart_fit_all_5perc <-
HvC_Heart_fit_all[which(HvC_Heart_fit_all$adj.P.Val < FDR_level), ]
human_chimp_heart1 <- rownames(methyl) %in% rownames( HvC_Heart_fit_all_5perc)
human_chimp_heart1 <- as.data.frame(human_chimp_heart1)
counts_genes_in <- cbind(methyl, human_chimp_heart1)
counts_genes_in_cutoff <- subset(counts_genes_in, human_chimp_heart1 == "TRUE")
counts_human_chimp_heart_subset <- counts_genes_in_cutoff[,1:7]
M <- counts_human_chimp_heart_subset
human_chimp_heart1 <- rownames(hc_exprs_heart) %in% rownames( HvC_Heart_fit_all_5perc)
human_chimp_heart1 <- as.data.frame(human_chimp_heart1)
counts_genes_in <- cbind(hc_exprs_heart, human_chimp_heart1)
counts_genes_in_cutoff <- subset(counts_genes_in, human_chimp_heart1 == "TRUE")
counts_human_chimp_heart_subset <- counts_genes_in_cutoff[,1:7]
Y <- counts_human_chimp_heart_subset
Human heart versus kidney- conserved DE genes
# Species no DE
species_no_DE <- c(32, 33, 36, 37, 40, 41, 44, 45)
species_no_DE_exprs <- hc_exprs[,species_no_DE]
species_no_DE_methyl <- exprs_methyl[,species_no_DE]
two_species <- samples$Tissue[species_no_DE]
no_DE_tissue <- droplevels.factor(two_species)
no_DE_RIN <- RIN[species_no_DE]
design <- model.matrix(~ as.factor(no_DE_tissue) + no_DE_RIN)
fit_species_no_DE <- lmFit(cpm.voom.cyclic[,species_no_DE], design)
fit_species_no_DE <- eBayes(fit_species_no_DE)
HvC_Heart_fit_species_no_DE = topTable(fit_species_no_DE, coef=2, adjust="BH", number=Inf,
sort.by="none")
# Species DE
design_1 <- model.matrix(~ X + RIN_subset)
fit_all <- lmFit(cpm.voom.cyclic[,human_chimp_heart], design_1)
fit_all <- eBayes(fit_all)
# Get results
HvC_Heart_fit_all = topTable(fit_all, coef=2, adjust="BH", number=Inf, sort.by="none")
HvC_Heart_fit_species_no_DE = cbind(HvC_Heart_fit_species_no_DE, HvC_Heart_fit_all)
HvC_Heart_fit_all_5perc <- HvC_Heart_fit_species_no_DE[which(HvC_Heart_fit_species_no_DE[,6] < FDR_level & HvC_Heart_fit_species_no_DE[,13] < FDR_level), ]
human_chimp_heart1 <- rownames(methyl) %in% rownames( HvC_Heart_fit_all_5perc)
human_chimp_heart1 <- as.data.frame(human_chimp_heart1)
counts_genes_in <- cbind(methyl, human_chimp_heart1)
counts_genes_in_cutoff <- subset(counts_genes_in, human_chimp_heart1 == "TRUE")
counts_human_chimp_heart_subset <- counts_genes_in_cutoff[,1:7]
M <- counts_human_chimp_heart_subset
human_chimp_heart1 <- rownames(hc_exprs_heart) %in% rownames( HvC_Heart_fit_all_5perc)
human_chimp_heart1 <- as.data.frame(human_chimp_heart1)
counts_genes_in <- cbind(hc_exprs_heart, human_chimp_heart1)
counts_genes_in_cutoff <- subset(counts_genes_in, human_chimp_heart1 == "TRUE")
counts_human_chimp_heart_subset <- counts_genes_in_cutoff[,1:7]
Y <- counts_human_chimp_heart_subset
# Save
cons_DE_heart_kidney_exp <- Y
cons_DE_heart_kidney_meth <- M
# Find the correlation
corr_cons_DE_HK <- array("NA", dim=c(nrow(Y),1))
for (i in 1:nrow(Y)){
corr_cons_DE_HK[i,] <- cor(t(cons_DE_heart_kidney_exp[i,]), t(cons_DE_heart_kidney_meth[i,]))
}
corr_cons_DE_HK <- as.numeric(corr_cons_DE_HK)
summary(corr_cons_DE_HK)
## Min. 1st Qu. Median Mean 3rd Qu. Max.
## -0.9923 -0.5504 -0.1663 -0.1252 0.2856 0.9713
Non conserved
# Species no DE
species_no_DE <- c(32, 33, 36, 37, 40, 41, 44, 45)
species_no_DE_exprs <- hc_exprs[,species_no_DE]
species_no_DE_methyl <- exprs_methyl[,species_no_DE]
two_species <- samples$Tissue[species_no_DE]
no_DE_tissue <- droplevels.factor(two_species)
no_DE_RIN <- RIN[species_no_DE]
design <- model.matrix(~ as.factor(no_DE_tissue) + no_DE_RIN)
fit_species_no_DE <- lmFit(cpm.voom.cyclic[,species_no_DE], design)
fit_species_no_DE <- eBayes(fit_species_no_DE)
HvC_Heart_fit_species_no_DE = topTable(fit_species_no_DE, coef=2, adjust="BH", number=Inf,
sort.by="none")
# Species DE
design_1 <- model.matrix(~ X + RIN_subset)
fit_all <- lmFit(cpm.voom.cyclic[,human_chimp_heart], design_1)
fit_all <- eBayes(fit_all)
# Get results
HvC_Heart_fit_all = topTable(fit_all, coef=2, adjust="BH", number=Inf, sort.by="none")
HvC_Heart_fit_species_no_DE = cbind(HvC_Heart_fit_species_no_DE, HvC_Heart_fit_all)
HvC_Heart_fit_all_5perc <- HvC_Heart_fit_species_no_DE[which(HvC_Heart_fit_species_no_DE[,6] > FDR_level & HvC_Heart_fit_species_no_DE[,13] < FDR_level), ]
human_chimp_heart1 <- rownames(methyl) %in% rownames( HvC_Heart_fit_all_5perc)
human_chimp_heart1 <- as.data.frame(human_chimp_heart1)
counts_genes_in <- cbind(methyl, human_chimp_heart1)
counts_genes_in_cutoff <- subset(counts_genes_in, human_chimp_heart1 == "TRUE")
counts_human_chimp_heart_subset <- counts_genes_in_cutoff[,1:7]
M <- counts_human_chimp_heart_subset
human_chimp_heart1 <- rownames(hc_exprs_heart) %in% rownames( HvC_Heart_fit_all_5perc)
human_chimp_heart1 <- as.data.frame(human_chimp_heart1)
counts_genes_in <- cbind(hc_exprs_heart, human_chimp_heart1)
counts_genes_in_cutoff <- subset(counts_genes_in, human_chimp_heart1 == "TRUE")
counts_human_chimp_heart_subset <- counts_genes_in_cutoff[,1:7]
Y <- counts_human_chimp_heart_subset
non_cons_DE_heart_kidney_exp <- Y
non_cons_DE_heart_kidney_meth <- M
# Find the correlation
corr_non_cons_DE_HK <- array("NA", dim=c(nrow(Y),1))
for (i in 1:nrow(Y)){
corr_non_cons_DE_HK[i,] <- cor(t(non_cons_DE_heart_kidney_exp[i,]), t(non_cons_DE_heart_kidney_meth[i,]))
}
corr_non_cons_DE_HK <- as.numeric(corr_non_cons_DE_HK)
summary(corr_non_cons_DE_HK)
## Min. 1st Qu. Median Mean 3rd Qu. Max.
## -0.93351 -0.46632 -0.12835 -0.09711 0.19282 0.93242
Human heart versus kidney- non DE genes
# Run the linear model in limma
design_1 <- model.matrix(~ X + RIN_subset)
fit_all <- lmFit(cpm.voom.cyclic[,human_chimp_heart], design_1)
fit_all <- eBayes(fit_all)
# Get results
HvC_Heart_fit_all = topTable(fit_all, coef=2, adjust="BH", number=Inf, sort.by="none")
HvC_Heart_fit_all_5perc <-
HvC_Heart_fit_all[which(HvC_Heart_fit_all$adj.P.Val > FDR_level), ]
human_chimp_heart1 <- rownames(methyl) %in% rownames( HvC_Heart_fit_all_5perc)
human_chimp_heart1 <- as.data.frame(human_chimp_heart1)
counts_genes_in <- cbind(methyl, human_chimp_heart1)
counts_genes_in_cutoff <- subset(counts_genes_in, human_chimp_heart1 == "TRUE")
counts_human_chimp_heart_subset <- counts_genes_in_cutoff[,1:7]
M <- counts_human_chimp_heart_subset
human_chimp_heart1 <- rownames(hc_exprs_heart) %in% rownames( HvC_Heart_fit_all_5perc)
human_chimp_heart1 <- as.data.frame(human_chimp_heart1)
counts_genes_in <- cbind(hc_exprs_heart, human_chimp_heart1)
counts_genes_in_cutoff <- subset(counts_genes_in, human_chimp_heart1 == "TRUE")
counts_human_chimp_heart_subset <- counts_genes_in_cutoff[,1:7]
Y <- counts_human_chimp_heart_subset
non_DE_heart_kidney_exp <- Y
non_DE_heart_kidney_meth <- M
# Find the correlation
corr_non_DE_HK <- array("NA", dim=c(nrow(Y),1))
for (i in 1:nrow(Y)){
corr_non_DE_HK[i,] <- cor(t(non_DE_heart_kidney_exp[i,]), t(non_DE_heart_kidney_meth[i,]))
}
corr_non_DE_HK <- as.numeric(corr_non_DE_HK)
summary(corr_non_DE_HK)
## Min. 1st Qu. Median Mean 3rd Qu. Max.
## -0.96488 -0.35283 -0.03606 -0.02938 0.29530 0.95263
Attempt to make this into a figure
length(corr_cons_DE_HK)
## [1] 689
length(corr_non_cons_DE_HK)
## [1] 97
length(corr_non_DE_HK)
## [1] 3369
corr_cons_DE_HK <- as.data.frame(corr_cons_DE_HK)
cons_label <- array("Conserved DE", dim=c(nrow(corr_cons_DE_HK),1))
corr_cons_DE_HK_full <- as.data.frame(cbind(corr_cons_DE_HK, cons_label))
colnames(corr_cons_DE_HK_full) <- c("Pearson's correlation", "DE status")
corr_non_cons_DE_HK <- as.data.frame(corr_non_cons_DE_HK)
non_cons_label <- array("Human only DE", dim=c(nrow(corr_non_cons_DE_HK),1))
corr_non_cons_DE_HK_full <- as.data.frame(cbind(corr_non_cons_DE_HK, non_cons_label))
colnames(corr_non_cons_DE_HK_full) <- c("Pearson's correlation", "DE status")
corr_non_DE_HK <- as.data.frame(corr_non_DE_HK)
non_de_label <- array("Human non-DE", dim=c(nrow(corr_non_DE_HK),1))
corr_non_DE_HK_full <- as.data.frame(cbind(corr_non_DE_HK, non_de_label))
colnames(corr_non_DE_HK_full) <- c("Pearson's correlation", "DE status")
# Make correlation
human_HK <- rbind(corr_cons_DE_HK_full, corr_non_cons_DE_HK_full, corr_non_DE_HK_full)
# Make mean
mean_cons <- mean(t(corr_cons_DE_HK))
mean_cons_label <- array("Conserved DE", dim=c(1,1))
mean_cons_full <- cbind(mean_cons, mean_cons_label)
colnames(mean_cons_full) <- c("Mean", "DE status")
mean_non_cons <- mean(t(corr_non_cons_DE_HK))
mean_non_cons_label <- array("Human only DE", dim=c(1,1))
mean_non_cons_full <- cbind(mean_non_cons, mean_non_cons_label)
colnames(mean_non_cons_full) <- c("Mean", "DE status")
mean_non_de <- mean(t(corr_non_DE_HK))
mean_non_de_label <- array("Human non-DE", dim=c(1,1))
mean_non_de_full <- cbind(mean_non_de, mean_non_de_label)
colnames(mean_non_de_full) <- c("Mean", "DE status")
all_mean <- as.data.frame(rbind(mean_cons_full, mean_non_cons_full, mean_non_de_full), stringsAsFactors = FALSE)
all_mean[,1] <- as.numeric(all_mean[,1])
# Plot without mean
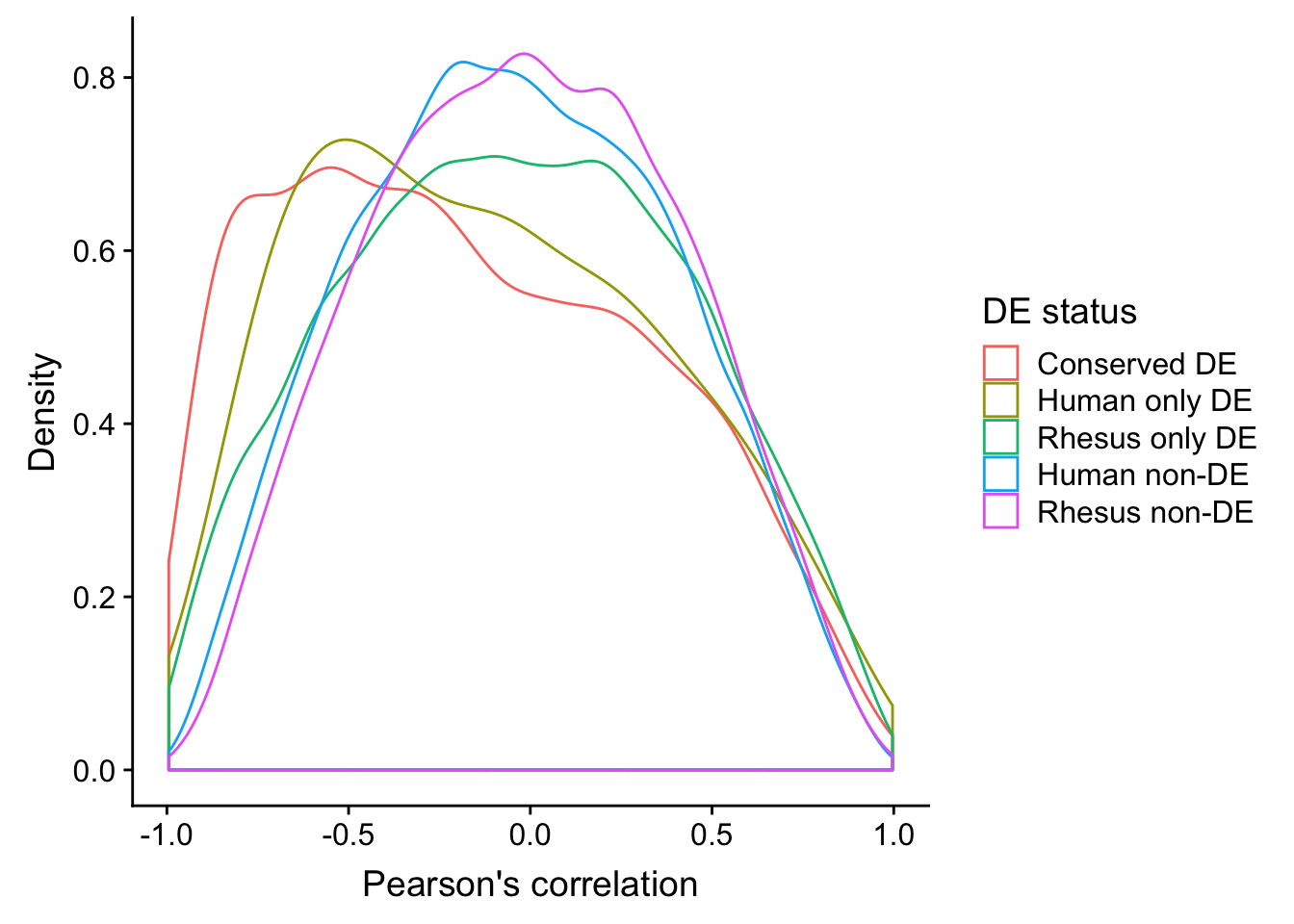
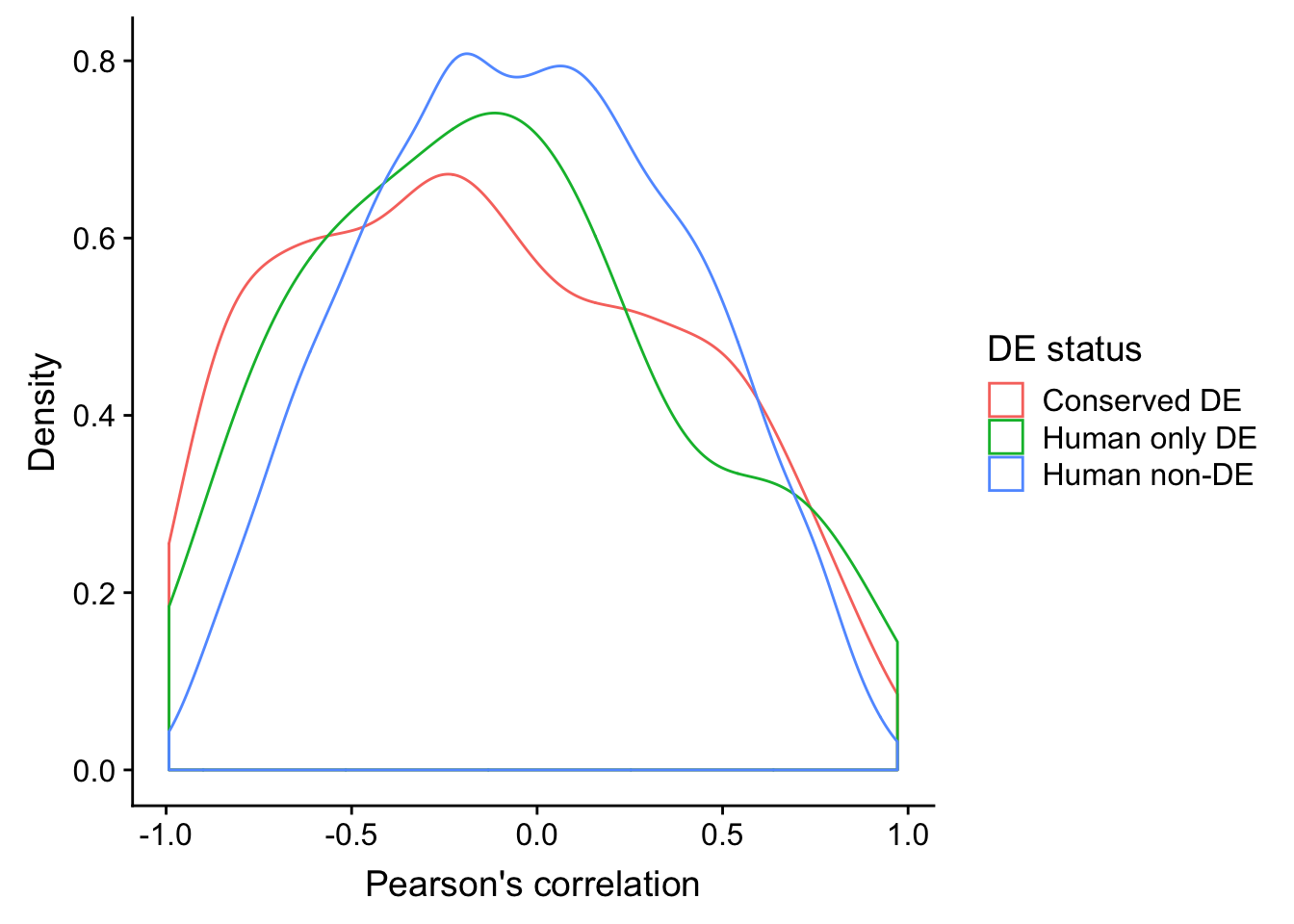
ggplot(human_HK, aes(x=`Pearson's correlation`, color=`DE status`)) +
geom_density() + ylab("Density")
# Plot with mean
#ggplot(human_HK, aes(x=`Pearson's correlation`, color=`DE status`)) +
# geom_density() + geom_vline(data=all_mean, aes(xintercept=Mean, # color=`DE status`), linetype="dashed")
Generalize conserved DE to a function
cons_de_corr <- function(human_chimp_heart, species_no_DE){
human_chimp_heart <- human_chimp_heart
# Methylation
# Methylation
hc_exprs <- exprs[,2:48]
exprs_methyl <- exprs[,49:95]
# HC heart only
hc_exprs_heart <- hc_exprs[,human_chimp_heart]
methyl <- exprs_methyl[,human_chimp_heart]
rownames(hc_exprs_heart) <- exprs[,1]
rownames(methyl) <- exprs[,1]
Y <- hc_exprs_heart
two_species <- samples$Tissue[human_chimp_heart]
X <- droplevels.factor(two_species)
M <- methyl
RIN_subset <- RIN[human_chimp_heart]
## Human heart versus kidney- DE genes
# Run the linear model in limma
design_1 <- model.matrix(~ X + RIN_subset)
fit_all <- lmFit(cpm.voom.cyclic[,human_chimp_heart], design_1)
fit_all <- eBayes(fit_all)
# Get results
HvC_Heart_fit_all = topTable(fit_all, coef=2, adjust="BH", number=Inf, sort.by="none")
HvC_Heart_fit_all_5perc <-
HvC_Heart_fit_all[which(HvC_Heart_fit_all$adj.P.Val < FDR_level), ]
human_chimp_heart1 <- rownames(methyl) %in% rownames( HvC_Heart_fit_all_5perc)
human_chimp_heart1 <- as.data.frame(human_chimp_heart1)
counts_genes_in <- cbind(methyl, human_chimp_heart1)
counts_genes_in_cutoff <- subset(counts_genes_in, human_chimp_heart1 == "TRUE")
counts_human_chimp_heart_subset <- counts_genes_in_cutoff[,1:7]
M <- counts_human_chimp_heart_subset
human_chimp_heart1 <- rownames(hc_exprs_heart) %in% rownames( HvC_Heart_fit_all_5perc)
human_chimp_heart1 <- as.data.frame(human_chimp_heart1)
counts_genes_in <- cbind(hc_exprs_heart, human_chimp_heart1)
counts_genes_in_cutoff <- subset(counts_genes_in, human_chimp_heart1 == "TRUE")
counts_human_chimp_heart_subset <- counts_genes_in_cutoff[,1:7]
Y <- counts_human_chimp_heart_subset
## Human heart versus kidney- conserved DE genes
# Species no DE
species_no_DE <- species_no_DE
species_no_DE_exprs <- hc_exprs[,species_no_DE]
species_no_DE_methyl <- exprs_methyl[,species_no_DE]
two_species <- samples$Tissue[species_no_DE]
no_DE_tissue <- droplevels.factor(two_species)
no_DE_RIN <- RIN[species_no_DE]
design <- model.matrix(~ as.factor(no_DE_tissue) + no_DE_RIN)
fit_species_no_DE <- lmFit(cpm.voom.cyclic[,species_no_DE], design)
fit_species_no_DE <- eBayes(fit_species_no_DE)
HvC_Heart_fit_species_no_DE = topTable(fit_species_no_DE, coef=2, adjust="BH", number=Inf,
sort.by="none")
# Species DE
design_1 <- model.matrix(~ X + RIN_subset)
fit_all <- lmFit(cpm.voom.cyclic[,human_chimp_heart], design_1)
fit_all <- eBayes(fit_all)
# Get results
HvC_Heart_fit_all = topTable(fit_all, coef=2, adjust="BH", number=Inf, sort.by="none")
HvC_Heart_fit_species_no_DE = cbind(HvC_Heart_fit_species_no_DE, HvC_Heart_fit_all)
HvC_Heart_fit_all_5perc <- HvC_Heart_fit_species_no_DE[which(HvC_Heart_fit_species_no_DE[,6] < FDR_level & HvC_Heart_fit_species_no_DE[,13] < FDR_level), ]
human_chimp_heart1 <- rownames(methyl) %in% rownames( HvC_Heart_fit_all_5perc)
human_chimp_heart1 <- as.data.frame(human_chimp_heart1)
counts_genes_in <- cbind(methyl, human_chimp_heart1)
counts_genes_in_cutoff <- subset(counts_genes_in, human_chimp_heart1 == "TRUE")
counts_human_chimp_heart_subset <- counts_genes_in_cutoff[,1:7]
M <- counts_human_chimp_heart_subset
human_chimp_heart1 <- rownames(hc_exprs_heart) %in% rownames( HvC_Heart_fit_all_5perc)
human_chimp_heart1 <- as.data.frame(human_chimp_heart1)
counts_genes_in <- cbind(hc_exprs_heart, human_chimp_heart1)
counts_genes_in_cutoff <- subset(counts_genes_in, human_chimp_heart1 == "TRUE")
counts_human_chimp_heart_subset <- counts_genes_in_cutoff[,1:7]
Y <- counts_human_chimp_heart_subset
# Save
cons_DE_heart_kidney_exp <- Y
cons_DE_heart_kidney_meth <- M
# Find the correlation
corr_cons_DE_HK <- array("NA", dim=c(nrow(Y),1))
for (i in 1:nrow(Y)){
corr_cons_DE_HK[i,] <- cor(t(cons_DE_heart_kidney_exp[i,]), t(cons_DE_heart_kidney_meth[i,]))
}
corr_cons_DE_HK <- as.numeric(corr_cons_DE_HK)
return(corr_cons_DE_HK)
}
# Heart kidney
hc_heart_kidney <- as.data.frame(cons_de_corr(c(17, 20, 21, 24, 25, 28, 29),c(32, 33, 36, 37, 40, 41, 44, 45)))
colnames(hc_heart_kidney) <- c("Pearson's correlation")
summary(corr_cons_DE_HK)
## corr_cons_DE_HK
## Min. :-0.9923
## 1st Qu.:-0.5504
## Median :-0.1663
## Mean :-0.1252
## 3rd Qu.: 0.2856
## Max. : 0.9713
summary(hc_heart_kidney)
## Pearson's correlation
## Min. :-0.9923
## 1st Qu.:-0.5504
## Median :-0.1663
## Mean :-0.1252
## 3rd Qu.: 0.2856
## Max. : 0.9713
# Heart liver
hc_heart_liver <- as.data.frame(cons_de_corr(c(18, 20, 22, 24, 26, 28, 30),c(32, 34, 36, 38, 40, 42, 44, 46)))
colnames(hc_heart_liver) <- c("Pearson's correlation")
summary(hc_heart_liver)
## Pearson's correlation
## Min. :-0.9716
## 1st Qu.:-0.6579
## Median :-0.3086
## Mean :-0.2264
## 3rd Qu.: 0.1709
## Max. : 0.9892
# Heart lung
hc_heart_lung <- as.data.frame(cons_de_corr(c(19, 20, 23, 24, 27, 28, 31),c(32, 35, 36, 39, 40, 43, 44, 47)))
colnames(hc_heart_lung) <- c("Pearson's correlation")
summary(hc_heart_lung)
## Pearson's correlation
## Min. :-0.9680
## 1st Qu.:-0.5175
## Median :-0.1485
## Mean :-0.1164
## 3rd Qu.: 0.2729
## Max. : 0.9264
# Kidney liver
hc_kidney_liver <- as.data.frame(cons_de_corr(c(17, 18, 21, 22, 25, 26, 29, 30),c(33, 34, 37, 38, 41, 42, 45, 46)))
colnames(hc_kidney_liver) <- c("Pearson's correlation")
summary(hc_kidney_liver)
## Pearson's correlation
## Min. :-0.9797
## 1st Qu.:-0.5773
## Median :-0.2038
## Mean :-0.1697
## 3rd Qu.: 0.2144
## Max. : 0.9746
# Kidney lung
hc_kidney_lung <- as.data.frame(cons_de_corr(c(17, 19, 21, 23, 25, 27, 29, 31),c(33, 35, 37, 39, 41, 43, 45, 47)))
colnames(hc_kidney_lung) <- c("Pearson's correlation")
summary(hc_kidney_lung)
## Pearson's correlation
## Min. :-0.9948
## 1st Qu.:-0.6056
## Median :-0.2333
## Mean :-0.1843
## 3rd Qu.: 0.2154
## Max. : 0.9530
# Liver lung
hc_liver_lung <- as.data.frame(cons_de_corr(c(18, 19, 22, 23, 26, 27, 30, 31),c(34, 35, 38, 39, 42, 43, 46, 47)))
colnames(hc_liver_lung) <- c("Pearson's correlation")
summary(hc_liver_lung)
## Pearson's correlation
## Min. :-0.9881
## 1st Qu.:-0.6416
## Median :-0.3026
## Mean :-0.2183
## 3rd Qu.: 0.1693
## Max. : 0.9408
hc_cons_de <- rbind(hc_heart_kidney, hc_heart_liver, hc_heart_lung, hc_kidney_liver, hc_kidney_lung, hc_liver_lung)
dim(hc_cons_de)
## [1] 3702 1
#hist(hc_cons_de)
write.table(hc_cons_de, "/Users/laurenblake/Desktop/Regulatory_Evol/ashlar-trial/data/Corr_human_rhesus_DE/corr_hr_de.txt")
Generalize species only DE to a function
non_cons_de_corr <- function(human_chimp_heart, species_no_DE){
human_chimp_heart <- human_chimp_heart
# Methylation
hc_exprs <- exprs[,2:48]
exprs_methyl <- exprs[,49:95]
# HC heart only
hc_exprs_heart <- hc_exprs[,human_chimp_heart]
methyl <- exprs_methyl[,human_chimp_heart]
rownames(hc_exprs_heart) <- exprs[,1]
rownames(methyl) <- exprs[,1]
Y <- hc_exprs_heart
two_species <- samples$Tissue[human_chimp_heart]
X <- droplevels.factor(two_species)
M <- methyl
RIN_subset <- RIN[human_chimp_heart]
## Human heart versus kidney- DE genes
# Run the linear model in limma
design_1 <- model.matrix(~ X + RIN_subset)
fit_all <- lmFit(cpm.voom.cyclic[,human_chimp_heart], design_1)
fit_all <- eBayes(fit_all)
# Get results
HvC_Heart_fit_all = topTable(fit_all, coef=2, adjust="BH", number=Inf, sort.by="none")
HvC_Heart_fit_all_5perc <-
HvC_Heart_fit_all[which(HvC_Heart_fit_all$adj.P.Val < FDR_level), ]
human_chimp_heart1 <- rownames(methyl) %in% rownames( HvC_Heart_fit_all_5perc)
human_chimp_heart1 <- as.data.frame(human_chimp_heart1)
counts_genes_in <- cbind(methyl, human_chimp_heart1)
counts_genes_in_cutoff <- subset(counts_genes_in, human_chimp_heart1 == "TRUE")
counts_human_chimp_heart_subset <- counts_genes_in_cutoff[,1:7]
M <- counts_human_chimp_heart_subset
human_chimp_heart1 <- rownames(hc_exprs_heart) %in% rownames( HvC_Heart_fit_all_5perc)
human_chimp_heart1 <- as.data.frame(human_chimp_heart1)
counts_genes_in <- cbind(hc_exprs_heart, human_chimp_heart1)
counts_genes_in_cutoff <- subset(counts_genes_in, human_chimp_heart1 == "TRUE")
counts_human_chimp_heart_subset <- counts_genes_in_cutoff[,1:7]
Y <- counts_human_chimp_heart_subset
## Human heart versus kidney- conserved DE genes
# Species no DE
species_no_DE <- species_no_DE
species_no_DE_exprs <- hc_exprs[,species_no_DE]
species_no_DE_methyl <- exprs_methyl[,species_no_DE]
two_species <- samples$Tissue[species_no_DE]
no_DE_tissue <- droplevels.factor(two_species)
no_DE_RIN <- RIN[species_no_DE]
design <- model.matrix(~ as.factor(no_DE_tissue) + no_DE_RIN)
fit_species_no_DE <- lmFit(cpm.voom.cyclic[,species_no_DE], design)
fit_species_no_DE <- eBayes(fit_species_no_DE)
HvC_Heart_fit_species_no_DE = topTable(fit_species_no_DE, coef=2, adjust="BH", number=Inf,
sort.by="none")
# Species DE
design_1 <- model.matrix(~ X + RIN_subset)
fit_all <- lmFit(cpm.voom.cyclic[,human_chimp_heart], design_1)
fit_all <- eBayes(fit_all)
# Get results
HvC_Heart_fit_all = topTable(fit_all, coef=2, adjust="BH", number=Inf, sort.by="none")
HvC_Heart_fit_species_no_DE = cbind(HvC_Heart_fit_species_no_DE, HvC_Heart_fit_all)
HvC_Heart_fit_all_5perc <- HvC_Heart_fit_species_no_DE[which(HvC_Heart_fit_species_no_DE[,6] > FDR_level & HvC_Heart_fit_species_no_DE[,13] < FDR_level), ]
human_chimp_heart1 <- rownames(methyl) %in% rownames( HvC_Heart_fit_all_5perc)
human_chimp_heart1 <- as.data.frame(human_chimp_heart1)
counts_genes_in <- cbind(methyl, human_chimp_heart1)
counts_genes_in_cutoff <- subset(counts_genes_in, human_chimp_heart1 == "TRUE")
counts_human_chimp_heart_subset <- counts_genes_in_cutoff[,1:7]
M <- counts_human_chimp_heart_subset
human_chimp_heart1 <- rownames(hc_exprs_heart) %in% rownames( HvC_Heart_fit_all_5perc)
human_chimp_heart1 <- as.data.frame(human_chimp_heart1)
counts_genes_in <- cbind(hc_exprs_heart, human_chimp_heart1)
counts_genes_in_cutoff <- subset(counts_genes_in, human_chimp_heart1 == "TRUE")
counts_human_chimp_heart_subset <- counts_genes_in_cutoff[,1:7]
Y <- counts_human_chimp_heart_subset
# Save
cons_DE_heart_kidney_exp <- Y
cons_DE_heart_kidney_meth <- M
# Find the correlation
corr_cons_DE_HK <- array("NA", dim=c(nrow(Y),1))
for (i in 1:nrow(Y)){
corr_cons_DE_HK[i,] <- cor(t(cons_DE_heart_kidney_exp[i,]), t(cons_DE_heart_kidney_meth[i,]))
}
corr_cons_DE_HK <- as.numeric(corr_cons_DE_HK)
return(corr_cons_DE_HK)
}
# Heart kidney
hc_heart_kidney <- as.data.frame(non_cons_de_corr(c(17, 20, 21, 24, 25, 28, 29),c(32, 33, 36, 37, 40, 41, 44, 45)))
colnames(hc_heart_kidney) <- c("Pearson's correlation")
summary(corr_non_cons_DE_HK)
## corr_non_cons_DE_HK
## Min. :-0.93351
## 1st Qu.:-0.46632
## Median :-0.12835
## Mean :-0.09711
## 3rd Qu.: 0.19282
## Max. : 0.93242
summary(hc_heart_kidney)
## Pearson's correlation
## Min. :-0.93351
## 1st Qu.:-0.46632
## Median :-0.12835
## Mean :-0.09711
## 3rd Qu.: 0.19282
## Max. : 0.93242
# Heart liver
hc_heart_liver <- as.data.frame(non_cons_de_corr(c(18, 20, 22, 24, 26, 28, 30),c(32, 34, 36, 38, 40, 42, 44, 46)))
colnames(hc_heart_liver) <- c("Pearson's correlation")
summary(hc_heart_liver)
## Pearson's correlation
## Min. :-0.9296
## 1st Qu.:-0.5804
## Median :-0.2690
## Mean :-0.1926
## 3rd Qu.: 0.2013
## Max. : 0.9706
# Heart lung
hc_heart_lung <- as.data.frame(non_cons_de_corr(c(19, 20, 23, 24, 27, 28, 31),c(32, 35, 36, 39, 40, 43, 44, 47)))
colnames(hc_heart_lung) <- c("Pearson's correlation")
summary(hc_heart_lung)
## Pearson's correlation
## Min. :-0.81110
## 1st Qu.:-0.41416
## Median :-0.08021
## Mean :-0.06266
## 3rd Qu.: 0.27287
## Max. : 0.90132
# Kidney liver
hc_kidney_liver <- as.data.frame(non_cons_de_corr(c(17, 18, 21, 22, 25, 26, 29, 30),c(33, 34, 37, 38, 41, 42, 45, 46)))
colnames(hc_kidney_liver) <- c("Pearson's correlation")
summary(hc_kidney_liver)
## Pearson's correlation
## Min. :-0.97573
## 1st Qu.:-0.51345
## Median :-0.11801
## Mean :-0.08781
## 3rd Qu.: 0.30413
## Max. : 0.95867
# Kidney lung
hc_kidney_lung <- as.data.frame(non_cons_de_corr(c(17, 19, 21, 23, 25, 27, 29, 31),c(33, 35, 37, 39, 41, 43, 45, 47)))
colnames(hc_kidney_lung) <- c("Pearson's correlation")
summary(hc_kidney_lung)
## Pearson's correlation
## Min. :-0.8680
## 1st Qu.:-0.5829
## Median :-0.1320
## Mean :-0.1213
## 3rd Qu.: 0.2733
## Max. : 0.8605
# Liver lung
hc_liver_lung <- as.data.frame(non_cons_de_corr(c(18, 19, 22, 23, 26, 27, 30, 31),c(34, 35, 38, 39, 42, 43, 46, 47)))
colnames(hc_liver_lung) <- c("Pearson's correlation")
summary(hc_liver_lung)
## Pearson's correlation
## Min. :-0.90242
## 1st Qu.:-0.47890
## Median :-0.16139
## Mean :-0.09963
## 3rd Qu.: 0.25236
## Max. : 0.84300
non_cons_de_corr_human <- rbind(hc_heart_kidney, hc_heart_liver, hc_heart_lung, hc_kidney_liver, hc_kidney_lung, hc_liver_lung)
#hist(non_cons_de_corr_human)
dim(non_cons_de_corr_human)
## [1] 511 1
write.table(non_cons_de_corr_human, "/Users/laurenblake/Desktop/Regulatory_Evol/ashlar-trial/data/Corr_human_rhesus_DE/corr_hc_human_only_de.txt")
Rhesus DE only
# Heart kidney
hc_heart_kidney <- as.data.frame(non_cons_de_corr(c(32, 33, 36, 37, 40, 41, 44, 45),c(17, 20, 21, 24, 25, 28, 29)))
colnames(hc_heart_kidney) <- c("Pearson's correlation")
summary(corr_non_cons_DE_HK)
## corr_non_cons_DE_HK
## Min. :-0.93351
## 1st Qu.:-0.46632
## Median :-0.12835
## Mean :-0.09711
## 3rd Qu.: 0.19282
## Max. : 0.93242
summary(hc_heart_kidney)
## Pearson's correlation
## Min. :-0.98160
## 1st Qu.:-0.37764
## Median :-0.03355
## Mean :-0.02951
## 3rd Qu.: 0.31770
## Max. : 0.96815
# Heart liver
hc_heart_liver <- as.data.frame(non_cons_de_corr(c(32, 34, 36, 38, 40, 42, 44, 46), c(18, 20, 22, 24, 26, 28, 30)))
colnames(hc_heart_liver) <- c("Pearson's correlation")
summary(hc_heart_liver)
## Pearson's correlation
## Min. :-0.99367
## 1st Qu.:-0.45648
## Median :-0.06060
## Mean :-0.05882
## 3rd Qu.: 0.32987
## Max. : 0.99587
# Heart lung
hc_heart_lung <- as.data.frame(non_cons_de_corr(c(32, 35, 36, 39, 40, 43, 44, 47), c(19, 20, 23, 24, 27, 28, 31)))
colnames(hc_heart_lung) <- c("Pearson's correlation")
summary(hc_heart_lung)
## Pearson's correlation
## Min. :-0.98043
## 1st Qu.:-0.37164
## Median :-0.01988
## Mean :-0.02309
## 3rd Qu.: 0.31031
## Max. : 0.95657
# Kidney liver
hc_kidney_liver <- as.data.frame(non_cons_de_corr(c(33, 34, 37, 38, 41, 42, 45, 46), c(17, 18, 21, 22, 25, 26, 29, 30)))
colnames(hc_kidney_liver) <- c("Pearson's correlation")
summary(hc_kidney_liver)
## Pearson's correlation
## Min. :-0.99130
## 1st Qu.:-0.41743
## Median :-0.03232
## Mean :-0.04381
## 3rd Qu.: 0.31750
## Max. : 0.97602
# Kidney lung
hc_kidney_lung <- as.data.frame(non_cons_de_corr(c(33, 35, 37, 39, 41, 43, 45, 47), c(17, 19, 21, 23, 25, 27, 29, 31)))
colnames(hc_kidney_lung) <- c("Pearson's correlation")
summary(hc_kidney_lung)
## Pearson's correlation
## Min. :-0.98459
## 1st Qu.:-0.37084
## Median :-0.02116
## Mean :-0.02729
## 3rd Qu.: 0.32502
## Max. : 0.94720
# Liver lung
hc_liver_lung <- as.data.frame(non_cons_de_corr(c(34, 35, 38, 39, 42, 43, 46, 47), c(18, 19, 22, 23, 26, 27, 30, 31)))
colnames(hc_liver_lung) <- c("Pearson's correlation")
summary(hc_liver_lung)
## Pearson's correlation
## Min. :-0.98194
## 1st Qu.:-0.43327
## Median :-0.07833
## Mean :-0.05842
## 3rd Qu.: 0.31901
## Max. : 0.98662
non_cons_de_corr_chimp <- rbind(hc_heart_kidney, hc_heart_liver, hc_heart_lung, hc_kidney_liver, hc_kidney_lung, hc_liver_lung)
#hist(non_cons_de_corr_chimp)
dim(non_cons_de_corr_chimp)
## [1] 12960 1
write.table(non_cons_de_corr_chimp, "/Users/laurenblake/Desktop/Regulatory_Evol/ashlar-trial/data/Corr_human_rhesus_DE/corr_hc_rhesus_only_de.txt")
Genearalize non DE to a function
non_de_corr <- function(human_chimp_heart){
# Methylation
hc_exprs <- exprs[,2:48]
exprs_methyl <- exprs[,49:95]
# HC heart only
hc_exprs_heart <- hc_exprs[,human_chimp_heart]
methyl <- exprs_methyl[,human_chimp_heart]
rownames(hc_exprs_heart) <- exprs[,1]
rownames(methyl) <- exprs[,1]
Y <- hc_exprs_heart
two_species <- samples$Tissue[human_chimp_heart]
X <- droplevels.factor(two_species)
M <- methyl
RIN_subset <- RIN[human_chimp_heart]
# Run the linear model in limma
design_1 <- model.matrix(~ X + RIN_subset)
fit_all <- lmFit(cpm.voom.cyclic[,human_chimp_heart], design_1)
fit_all <- eBayes(fit_all)
# Get results
HvC_Heart_fit_all = topTable(fit_all, coef=2, adjust="BH", number=Inf, sort.by="none")
HvC_Heart_fit_all_5perc <-
HvC_Heart_fit_all[which(HvC_Heart_fit_all$adj.P.Val > FDR_level), ]
human_chimp_heart1 <- rownames(methyl) %in% rownames( HvC_Heart_fit_all_5perc)
human_chimp_heart1 <- as.data.frame(human_chimp_heart1)
counts_genes_in <- cbind(methyl, human_chimp_heart1)
counts_genes_in_cutoff <- subset(counts_genes_in, human_chimp_heart1 == "TRUE")
counts_human_chimp_heart_subset <- counts_genes_in_cutoff[,1:7]
M <- counts_human_chimp_heart_subset
human_chimp_heart1 <- rownames(hc_exprs_heart) %in% rownames( HvC_Heart_fit_all_5perc)
human_chimp_heart1 <- as.data.frame(human_chimp_heart1)
counts_genes_in <- cbind(hc_exprs_heart, human_chimp_heart1)
counts_genes_in_cutoff <- subset(counts_genes_in, human_chimp_heart1 == "TRUE")
counts_human_chimp_heart_subset <- counts_genes_in_cutoff[,1:7]
Y <- counts_human_chimp_heart_subset
non_DE_heart_kidney_exp <- Y
non_DE_heart_kidney_meth <- M
# Find the correlation
corr_non_DE_HK <- array("NA", dim=c(nrow(Y),1))
for (i in 1:nrow(Y)){
corr_non_DE_HK[i,] <- cor(t(non_DE_heart_kidney_exp[i,]), t(non_DE_heart_kidney_meth[i,]))
}
corr_non_DE_HK <- as.numeric(corr_non_DE_HK)
return(corr_non_DE_HK)
}
# Humans
# Heart kidney
hc_heart_kidney <- as.data.frame(non_de_corr(c(17, 20, 21, 24, 25, 28, 29)))
colnames(hc_heart_kidney) <- c("Pearson's correlation")
summary(corr_non_DE_HK)
## corr_non_DE_HK
## Min. :-0.96488
## 1st Qu.:-0.35283
## Median :-0.03606
## Mean :-0.02938
## 3rd Qu.: 0.29530
## Max. : 0.95263
summary(hc_heart_kidney)
## Pearson's correlation
## Min. :-0.96488
## 1st Qu.:-0.35283
## Median :-0.03606
## Mean :-0.02938
## 3rd Qu.: 0.29530
## Max. : 0.95263
# Heart liver
hc_heart_liver <- as.data.frame(non_de_corr(c(18, 20, 22, 24, 26, 28, 30)))
colnames(hc_heart_liver) <- c("Pearson's correlation")
summary(hc_heart_liver)
## Pearson's correlation
## Min. :-0.96733
## 1st Qu.:-0.38185
## Median :-0.06477
## Mean :-0.04895
## 3rd Qu.: 0.27933
## Max. : 0.94905
# Heart lung
hc_heart_lung <- as.data.frame(non_de_corr(c(19, 20, 23, 24, 27, 28, 31)))
colnames(hc_heart_lung) <- c("Pearson's correlation")
summary(hc_heart_lung)
## Pearson's correlation
## Min. :-0.96930
## 1st Qu.:-0.37019
## Median :-0.04352
## Mean :-0.03824
## 3rd Qu.: 0.28263
## Max. : 0.97460
# Kidney liver
hc_kidney_liver <- as.data.frame(non_de_corr(c(17, 18, 21, 22, 25, 26, 29, 30)))
colnames(hc_kidney_liver) <- c("Pearson's correlation")
summary(hc_kidney_liver)
## Pearson's correlation
## Min. :-0.98581
## 1st Qu.:-0.33652
## Median :-0.04219
## Mean :-0.03261
## 3rd Qu.: 0.26909
## Max. : 0.98979
# Kidney lung
hc_kidney_lung <- as.data.frame(non_de_corr(c(17, 19, 21, 23, 25, 27, 29, 31)))
colnames(hc_kidney_lung) <- c("Pearson's correlation")
summary(hc_kidney_lung)
## Pearson's correlation
## Min. :-0.98742
## 1st Qu.:-0.35645
## Median :-0.03271
## Mean :-0.02849
## 3rd Qu.: 0.30453
## Max. : 0.95967
# Liver lung
hc_liver_lung <- as.data.frame(non_de_corr(c(18, 19, 22, 23, 26, 27, 30, 31)))
colnames(hc_liver_lung) <- c("Pearson's correlation")
summary(hc_liver_lung)
## Pearson's correlation
## Min. :-0.97335
## 1st Qu.:-0.37339
## Median :-0.06036
## Mean :-0.04418
## 3rd Qu.: 0.28619
## Max. : 0.99670
hc_non_de_human <- rbind(hc_heart_kidney, hc_heart_liver, hc_heart_lung, hc_kidney_liver, hc_kidney_lung, hc_liver_lung)
dim(hc_non_de_human)
## [1] 20717 1
write.table(hc_non_de_human, "/Users/laurenblake/Desktop/Regulatory_Evol/ashlar-trial/data/Corr_human_rhesus_DE/corr_hc_human_nonde.txt")
# Chimps
# Heart kidney
hc_heart_kidney <- as.data.frame(non_de_corr(c(32, 33, 36, 37, 40, 41, 44, 45)))
colnames(hc_heart_kidney) <- c("Pearson's correlation")
summary(hc_heart_kidney)
## Pearson's correlation
## Min. :-0.965039
## 1st Qu.:-0.332512
## Median :-0.020129
## Mean :-0.006149
## 3rd Qu.: 0.309413
## Max. : 0.949220
# Heart liver
hc_heart_liver <- as.data.frame(non_de_corr(c(32, 34, 36, 38, 40, 42, 44, 46)))
colnames(hc_heart_liver) <- c("Pearson's correlation")
summary(hc_heart_liver)
## Pearson's correlation
## Min. :-0.934846
## 1st Qu.:-0.332182
## Median :-0.003170
## Mean : 0.001402
## 3rd Qu.: 0.340073
## Max. : 0.974967
# Heart lung
hc_heart_lung <- as.data.frame(non_de_corr(c(32, 35, 36, 39, 40, 43, 44, 47)))
colnames(hc_heart_lung) <- c("Pearson's correlation")
summary(hc_heart_lung)
## Pearson's correlation
## Min. :-0.946958
## 1st Qu.:-0.315109
## Median :-0.013413
## Mean :-0.009251
## 3rd Qu.: 0.295545
## Max. : 0.977170
# Kidney liver
hc_kidney_liver <- as.data.frame(non_de_corr(c(33, 34, 37, 38, 41, 42, 45, 46)))
colnames(hc_kidney_liver) <- c("Pearson's correlation")
summary(hc_kidney_liver)
## Pearson's correlation
## Min. :-0.974737
## 1st Qu.:-0.319052
## Median :-0.010582
## Mean :-0.004132
## 3rd Qu.: 0.299788
## Max. : 0.941500
# Kidney lung
hc_kidney_lung <- as.data.frame(non_de_corr(c(33, 35, 37, 39, 41, 43, 45, 47)))
colnames(hc_kidney_lung) <- c("Pearson's correlation")
summary(hc_kidney_lung)
## Pearson's correlation
## Min. :-0.92008
## 1st Qu.:-0.34419
## Median :-0.01823
## Mean :-0.01944
## 3rd Qu.: 0.30377
## Max. : 0.97514
# Liver lung
hc_liver_lung <- as.data.frame(non_de_corr(c(34, 35, 38, 39, 42, 43, 46, 47)))
colnames(hc_liver_lung) <- c("Pearson's correlation")
summary(hc_liver_lung)
## Pearson's correlation
## Min. :-0.95180
## 1st Qu.:-0.31987
## Median :-0.00467
## Mean :-0.01215
## 3rd Qu.: 0.28712
## Max. : 0.92538
hc_non_de_chimp <- rbind(hc_heart_kidney, hc_heart_liver, hc_heart_lung, hc_kidney_liver, hc_kidney_lung, hc_liver_lung)
dim(hc_non_de_chimp)
## [1] 8268 1
#hist(hc_non_de_chimp)
write.table(hc_non_de_chimp, "/Users/laurenblake/Desktop/Regulatory_Evol/ashlar-trial/data/Corr_human_rhesus_DE/corr_hc_rhesus_nonde.txt")