Identifying DMRs
Lauren Blake
January 5, 2018
Since identifying DMRs is computationally intensive using the BSmooth package, we needed to run this on the cluster (/mnt/gluster/home/leblake/Methylation/tissue_DMRs and /mnt/gluster/home/leblake/Methylation/species_DMRs)
First, we will examine the results of the tissue DMRs
# Import libraries
library("ggplot2")Warning: package 'ggplot2' was built under R version 3.4.4library("VennDiagram")Warning: package 'VennDiagram' was built under R version 3.4.4Loading required package: gridLoading required package: futile.loggerlibrary(plyr)
# Human tDMRs
humans_heart_kidney_DMRs <- read.delim("../data/humans_heart_kidney_DMRs.txt")
humans_heart_liver_DMRs <- read.delim("../data/humans_heart_liver_DMRs.txt")
humans_heart_lung_DMRs <- read.delim("../data/humans_heart_lung_DMRs.txt")
humans_liver_lung_DMRs <- read.delim("../data/humans_liver_lung_DMRs.txt")
humans_liver_kidney_DMRs <- read.delim("../data/humans_liver_kidney_DMRs.txt")
humans_lung_kidney_DMRs <- read.delim("../data/humans_lung_kidney_DMRs.txt")
levels(factor(humans_lung_kidney_DMRs[,1])) [1] "chr1" "chr10" "chr11" "chr12" "chr13" "chr14" "chr15" "chr16"
[9] "chr17" "chr18" "chr19" "chr2" "chr20" "chr21" "chr22" "chr3"
[17] "chr4" "chr5" "chr6" "chr7" "chr8" "chr9" "chrX" count(humans_lung_kidney_DMRs[,1]) x freq
1 chr1 1486
2 chr10 974
3 chr11 777
4 chr12 836
5 chr13 446
6 chr14 516
7 chr15 549
8 chr16 785
9 chr17 852
10 chr18 484
11 chr19 647
12 chr2 1280
13 chr20 506
14 chr21 284
15 chr22 412
16 chr3 765
17 chr4 746
18 chr5 799
19 chr6 850
20 chr7 992
21 chr8 777
22 chr9 758
23 chrX 158# Chimp tDMRs
chimps_heart_kidney_DMRs <- read.delim("../data/chimps_heart_kidney_DMRs.txt")
chimps_heart_liver_DMRs <- read.delim("../data/chimps_heart_liver_DMRs.txt")
chimps_heart_lung_DMRs <- read.delim("../data/chimps_heart_lung_DMRs.txt")
chimps_liver_lung_DMRs <- read.delim("../data/chimps_liver_lung_DMRs.txt")
chimps_liver_kidney_DMRs <- read.delim("../data/chimps_liver_kidney_DMRs.txt")
chimps_lung_kidney_DMRs <- read.delim("../data/chimps_lung_kidney_DMRs.txt")
levels(factor(chimps_lung_kidney_DMRs[,1])) [1] "chr1" "chr10"
[3] "chr11" "chr11_gl000202_random"
[5] "chr12" "chr13"
[7] "chr14" "chr15"
[9] "chr16" "chr17"
[11] "chr18" "chr19"
[13] "chr2" "chr20"
[15] "chr21" "chr22"
[17] "chr3" "chr4"
[19] "chr5" "chr6"
[21] "chr7" "chr8"
[23] "chr9" "chrX" count(chimps_lung_kidney_DMRs[,1]) x freq
1 chr1 1021
2 chr10 697
3 chr11 592
4 chr11_gl000202_random 1
5 chr12 621
6 chr13 327
7 chr14 408
8 chr15 396
9 chr16 528
10 chr17 587
11 chr18 348
12 chr19 375
13 chr2 1026
14 chr20 353
15 chr21 199
16 chr22 272
17 chr3 627
18 chr4 662
19 chr5 651
20 chr6 692
21 chr7 627
22 chr8 542
23 chr9 525
24 chrX 16# Rhesus tDMRs
rhesus_heart_kidney_DMRs <- read.delim("../data/rhesus_heart_kidney_DMRs.txt")
rhesus_heart_liver_DMRs <- read.delim("../data/rhesus_heart_liver_DMRs.txt")
rhesus_heart_lung_DMRs <- read.delim("../data/rhesus_heart_lung_DMRs.txt")
rhesus_liver_lung_DMRs <- read.delim("../data/rhesus_liver_lung_DMRs.txt")
rhesus_liver_kidney_DMRs <- read.delim("../data/rhesus_liver_kidney_DMRs.txt")
rhesus_lung_kidney_DMRs <- read.delim("../data/rhesus_lung_kidney_DMRs.txt")
levels(factor(rhesus_heart_kidney_DMRs[,1])) [1] "chr1" "chr10"
[3] "chr11" "chr11_gl000202_random"
[5] "chr12" "chr13"
[7] "chr14" "chr15"
[9] "chr16" "chr17"
[11] "chr17_gl000204_random" "chr18"
[13] "chr19" "chr2"
[15] "chr20" "chr21"
[17] "chr22" "chr3"
[19] "chr4" "chr5"
[21] "chr6" "chr7"
[23] "chr8" "chr9"
[25] "chrX" count(rhesus_lung_kidney_DMRs[,1]) x freq
1 chr1 1391
2 chr10 929
3 chr11 863
4 chr12 799
5 chr13 435
6 chr14 519
7 chr15 499
8 chr16 821
9 chr17 944
10 chr17_gl000204_random 1
11 chr18 412
12 chr19 655
13 chr2 1180
14 chr20 543
15 chr21 252
16 chr22 475
17 chr3 709
18 chr4 613
19 chr5 713
20 chr6 816
21 chr7 853
22 chr8 674
23 chr9 794
24 chrX 24How many regions overlap eachother?
library("bedr")
######################
#### bedr v1.0.4 ####
######################
checking binary availability...
* Checking path for bedtools... PASS
/usr/local/bin/bedtools
* Checking path for bedops... FAIL
* Checking path for tabix... FAIL
tests and examples will be skipped on R CMD check if binaries are missing# columns to pull
pull_col <- c(1:3, 16)
# make bed style format
humans_heart_liver_DMRs_bed <- humans_heart_liver_DMRs[,pull_col]
# Get DMRs on the autosomal chromosomes only
humans_heart_liver_DMRs_bed_noX <- humans_heart_liver_DMRs_bed[which(humans_heart_liver_DMRs_bed[,1] != "chrX"),]
humans_heart_liver_DMRs_bed_noX[,1] <- as.character(humans_heart_liver_DMRs_bed_noX[,1])
humans_heart_liver_DMRs_bed_noX[,2] <- as.integer(humans_heart_liver_DMRs_bed_noX[,2])
humans_heart_liver_DMRs_bed_noX[,3] <- as.integer(humans_heart_liver_DMRs_bed_noX[,3])
summary(humans_heart_liver_DMRs_bed_noX[,3] - humans_heart_liver_DMRs_bed_noX[,2]) Min. 1st Qu. Median Mean 3rd Qu. Max.
5.0 255.0 473.0 584.5 795.0 4797.0 sort_humans_heart_liver_DMRs_bed <- bedr.sort.region(humans_heart_liver_DMRs_bed_noX)SORTING
VALIDATE REGIONS
* Checking input type... PASS
Input is in bed format
* Check if index is a string... PASS
* Check index pattern... PASS
* Check for missing values... PASS
* Check for larger start position... PASS.
* Check if zero based... PASS
* Overlapping regions can cause unexpected results.# make bed style format
humans_heart_lung_DMRs_bed <- humans_heart_lung_DMRs[,pull_col]
humans_heart_lung_DMRs_bed_noX <- humans_heart_lung_DMRs_bed[which(humans_heart_lung_DMRs_bed[,1] != "chrX"),]
humans_heart_lung_DMRs_bed_noX[,1] <- as.character(humans_heart_lung_DMRs_bed_noX[,1])
humans_heart_lung_DMRs_bed_noX[,2] <- as.integer(humans_heart_lung_DMRs_bed_noX[,2])
humans_heart_lung_DMRs_bed_noX[,3] <- as.integer(humans_heart_lung_DMRs_bed_noX[,3])
sort_humans_heart_lung_DMRs_bed <- bedr.sort.region(humans_heart_lung_DMRs_bed_noX)SORTING
VALIDATE REGIONS
* Checking input type... PASS
Input is in bed format
* Check if index is a string... PASS
* Check index pattern... PASS
* Check for missing values... PASS
* Check for larger start position... PASS.
* Check if zero based... PASS
* Overlapping regions can cause unexpected results.summary(humans_heart_lung_DMRs_bed_noX[,3] - humans_heart_lung_DMRs_bed_noX[,2]) Min. 1st Qu. Median Mean 3rd Qu. Max.
4.0 225.0 419.0 516.6 713.0 3761.0 heart_lung <- bedr(input = list(a = sort_humans_heart_liver_DMRs_bed, b = sort_humans_heart_lung_DMRs_bed), method = "intersect", params = "-wao -f 0.5 -F 0.5") * Processing input (1): a
CONVERT TO BED
* Checking input type... PASS
Input is in bed format
VALIDATE REGIONS
* Check if index is a string... PASS
* Check index pattern... PASS
* Check for missing values... PASS
* Check for larger start position... PASS.
* Check if zero based... PASS
* Checking sort order... PASS
* Checking for overlapping 'contiguous' regions... PASS
* Processing input (2): b
CONVERT TO BED
* Checking input type... PASS
Input is in bed format
VALIDATE REGIONS
* Check if index is a string... PASS
* Check index pattern... PASS
* Check for missing values... PASS
* Check for larger start position... PASS.
* Check if zero based... PASS
* Checking sort order... PASS
* Checking for overlapping 'contiguous' regions... PASS
bedtools intersect -a /var/folders/rf/qrcw6ncj05z1pc_pq9xzw3540000gn/T//Rtmph6wW6Y/a_c3e33c087d10.bed -b /var/folders/rf/qrcw6ncj05z1pc_pq9xzw3540000gn/T//Rtmph6wW6Y/b_c3e32b054590.bed -wao -f 0.5 -F 0.5summary(heart_lung[,3] - heart_lung[,2]) Min. 1st Qu. Median Mean 3rd Qu. Max.
5.0 255.0 473.0 584.5 795.0 4797.0 summary(as.integer(heart_lung[,9])) Min. 1st Qu. Median Mean 3rd Qu. Max.
0.0 0.0 0.0 102.3 0.0 3167.0 summary(as.integer(heart_lung[,9]) > 0) Mode FALSE TRUE
logical 18703 3858 lung_heart <- bedr(input = list(a = sort_humans_heart_lung_DMRs_bed, b = sort_humans_heart_liver_DMRs_bed), method = "intersect", params = "-wao -f 0.5 -F 0.5") * Processing input (1): a
CONVERT TO BED
* Checking input type... PASS
Input is in bed format
VALIDATE REGIONS
* Check if index is a string... PASS
* Check index pattern... PASS
* Check for missing values... PASS
* Check for larger start position... PASS.
* Check if zero based... PASS
* Checking sort order... PASS
* Checking for overlapping 'contiguous' regions... PASS
* Processing input (2): b
CONVERT TO BED
* Checking input type... PASS
Input is in bed format
VALIDATE REGIONS
* Check if index is a string... PASS
* Check index pattern... PASS
* Check for missing values... PASS
* Check for larger start position... PASS.
* Check if zero based... PASS
* Checking sort order... PASS
* Checking for overlapping 'contiguous' regions... PASS
bedtools intersect -a /var/folders/rf/qrcw6ncj05z1pc_pq9xzw3540000gn/T//Rtmph6wW6Y/a_c3e35ca666dd.bed -b /var/folders/rf/qrcw6ncj05z1pc_pq9xzw3540000gn/T//Rtmph6wW6Y/b_c3e34266d7f.bed -wao -f 0.5 -F 0.5summary(as.integer(lung_heart[,9])) Min. 1st Qu. Median Mean 3rd Qu. Max.
0.0 0.0 0.0 162.5 162.0 3167.0 summary(as.integer(lung_heart[,9]) > 0) Mode FALSE TRUE
logical 10350 3858 # Make a Venn diagram based on the results
summary(as.integer(heart_lung[,9]) > 0) Mode FALSE TRUE
logical 18703 3858 summary(as.integer(lung_heart[,9]) > 0) Mode FALSE TRUE
logical 10350 3858 grid.newpage()
draw.pairwise.venn(area1 = 22561, area2 = 14208, cross.area = 3858, category = c("Heart v. Liver", "Heart v. Lung"))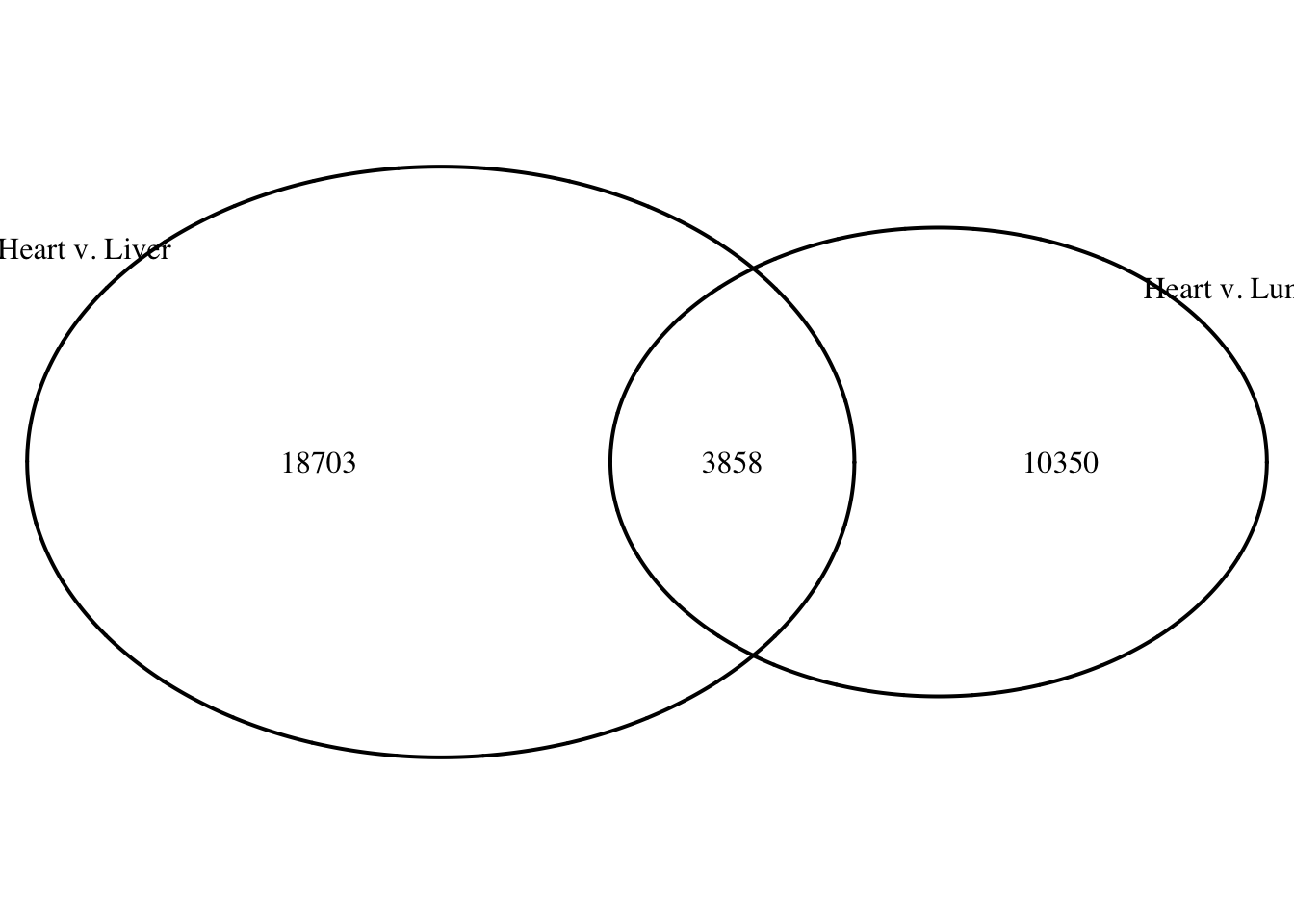
(polygon[GRID.polygon.1], polygon[GRID.polygon.2], polygon[GRID.polygon.3], polygon[GRID.polygon.4], text[GRID.text.5], text[GRID.text.6], text[GRID.text.7], text[GRID.text.8], text[GRID.text.9]) How does the overlap change based on % bp?
# 1 bp overlap
heart_liver_1 <- bedr(input = list(a = sort_humans_heart_liver_DMRs_bed, b = sort_humans_heart_lung_DMRs_bed), method = "intersect", params = "-wao -f 1E-9 -F 1E-9") * Processing input (1): a
CONVERT TO BED
* Checking input type... PASS
Input is in bed format
VALIDATE REGIONS
* Check if index is a string... PASS
* Check index pattern... PASS
* Check for missing values... PASS
* Check for larger start position... PASS.
* Check if zero based... PASS
* Checking sort order... PASS
* Checking for overlapping 'contiguous' regions... PASS
* Processing input (2): b
CONVERT TO BED
* Checking input type... PASS
Input is in bed format
VALIDATE REGIONS
* Check if index is a string... PASS
* Check index pattern... PASS
* Check for missing values... PASS
* Check for larger start position... PASS.
* Check if zero based... PASS
* Checking sort order... PASS
* Checking for overlapping 'contiguous' regions... PASS
bedtools intersect -a /var/folders/rf/qrcw6ncj05z1pc_pq9xzw3540000gn/T//Rtmph6wW6Y/a_c3e32e22332f.bed -b /var/folders/rf/qrcw6ncj05z1pc_pq9xzw3540000gn/T//Rtmph6wW6Y/b_c3e36ed5d06b.bed -wao -f 1E-9 -F 1E-9summary(as.integer(heart_liver_1[,9]) > 0) Mode FALSE TRUE
logical 16965 5607 # 10% overlap
heart_liver_10 <- bedr(input = list(a = sort_humans_heart_liver_DMRs_bed, b = sort_humans_heart_lung_DMRs_bed), method = "intersect", params = "-wao -f 0.10 -F 0.10") * Processing input (1): a
CONVERT TO BED
* Checking input type... PASS
Input is in bed format
VALIDATE REGIONS
* Check if index is a string... PASS
* Check index pattern... PASS
* Check for missing values... PASS
* Check for larger start position... PASS.
* Check if zero based... PASS
* Checking sort order... PASS
* Checking for overlapping 'contiguous' regions... PASS
* Processing input (2): b
CONVERT TO BED
* Checking input type... PASS
Input is in bed format
VALIDATE REGIONS
* Check if index is a string... PASS
* Check index pattern... PASS
* Check for missing values... PASS
* Check for larger start position... PASS.
* Check if zero based... PASS
* Checking sort order... PASS
* Checking for overlapping 'contiguous' regions... PASS
bedtools intersect -a /var/folders/rf/qrcw6ncj05z1pc_pq9xzw3540000gn/T//Rtmph6wW6Y/a_c3e3b81f65f.bed -b /var/folders/rf/qrcw6ncj05z1pc_pq9xzw3540000gn/T//Rtmph6wW6Y/b_c3e3594ca6a6.bed -wao -f 0.10 -F 0.10summary(as.integer(heart_liver_10[,9]) > 0) Mode FALSE TRUE
logical 17096 5472 # 20% overlap
heart_liver_20 <- bedr(input = list(a = sort_humans_heart_liver_DMRs_bed, b = sort_humans_heart_lung_DMRs_bed), method = "intersect", params = "-wao -f 0.20 -F 0.20") * Processing input (1): a
CONVERT TO BED
* Checking input type... PASS
Input is in bed format
VALIDATE REGIONS
* Check if index is a string... PASS
* Check index pattern... PASS
* Check for missing values... PASS
* Check for larger start position... PASS.
* Check if zero based... PASS
* Checking sort order... PASS
* Checking for overlapping 'contiguous' regions... PASS
* Processing input (2): b
CONVERT TO BED
* Checking input type... PASS
Input is in bed format
VALIDATE REGIONS
* Check if index is a string... PASS
* Check index pattern... PASS
* Check for missing values... PASS
* Check for larger start position... PASS.
* Check if zero based... PASS
* Checking sort order... PASS
* Checking for overlapping 'contiguous' regions... PASS
bedtools intersect -a /var/folders/rf/qrcw6ncj05z1pc_pq9xzw3540000gn/T//Rtmph6wW6Y/a_c3e356146bdf.bed -b /var/folders/rf/qrcw6ncj05z1pc_pq9xzw3540000gn/T//Rtmph6wW6Y/b_c3e32d3d1a5e.bed -wao -f 0.20 -F 0.20summary(as.integer(heart_liver_20[,9]) > 0) Mode FALSE TRUE
logical 17374 5189 # 25% overlap
heart_liver_25 <- bedr(input = list(a = sort_humans_heart_liver_DMRs_bed, b = sort_humans_heart_lung_DMRs_bed), method = "intersect", params = "-wao -f 0.25 -F 0.25") * Processing input (1): a
CONVERT TO BED
* Checking input type... PASS
Input is in bed format
VALIDATE REGIONS
* Check if index is a string... PASS
* Check index pattern... PASS
* Check for missing values... PASS
* Check for larger start position... PASS.
* Check if zero based... PASS
* Checking sort order... PASS
* Checking for overlapping 'contiguous' regions... PASS
* Processing input (2): b
CONVERT TO BED
* Checking input type... PASS
Input is in bed format
VALIDATE REGIONS
* Check if index is a string... PASS
* Check index pattern... PASS
* Check for missing values... PASS
* Check for larger start position... PASS.
* Check if zero based... PASS
* Checking sort order... PASS
* Checking for overlapping 'contiguous' regions... PASS
bedtools intersect -a /var/folders/rf/qrcw6ncj05z1pc_pq9xzw3540000gn/T//Rtmph6wW6Y/a_c3e321e33fe7.bed -b /var/folders/rf/qrcw6ncj05z1pc_pq9xzw3540000gn/T//Rtmph6wW6Y/b_c3e3504c6f64.bed -wao -f 0.25 -F 0.25summary(as.integer(heart_liver_25[,9]) > 0) Mode FALSE TRUE
logical 17554 5007 # 30% overlap
heart_liver_30 <- bedr(input = list(a = sort_humans_heart_liver_DMRs_bed, b = sort_humans_heart_lung_DMRs_bed), method = "intersect", params = "-wao -f 0.30 -F 0.30") * Processing input (1): a
CONVERT TO BED
* Checking input type... PASS
Input is in bed format
VALIDATE REGIONS
* Check if index is a string... PASS
* Check index pattern... PASS
* Check for missing values... PASS
* Check for larger start position... PASS.
* Check if zero based... PASS
* Checking sort order... PASS
* Checking for overlapping 'contiguous' regions... PASS
* Processing input (2): b
CONVERT TO BED
* Checking input type... PASS
Input is in bed format
VALIDATE REGIONS
* Check if index is a string... PASS
* Check index pattern... PASS
* Check for missing values... PASS
* Check for larger start position... PASS.
* Check if zero based... PASS
* Checking sort order... PASS
* Checking for overlapping 'contiguous' regions... PASS
bedtools intersect -a /var/folders/rf/qrcw6ncj05z1pc_pq9xzw3540000gn/T//Rtmph6wW6Y/a_c3e348ca2905.bed -b /var/folders/rf/qrcw6ncj05z1pc_pq9xzw3540000gn/T//Rtmph6wW6Y/b_c3e3707f3352.bed -wao -f 0.30 -F 0.30summary(as.integer(heart_liver_30[,9]) > 0) Mode FALSE TRUE
logical 17737 4824 #40% overlap
heart_liver_40 <- bedr(input = list(a = sort_humans_heart_liver_DMRs_bed, b = sort_humans_heart_lung_DMRs_bed), method = "intersect", params = "-wao -f 0.4 -F 0.4") * Processing input (1): a
CONVERT TO BED
* Checking input type... PASS
Input is in bed format
VALIDATE REGIONS
* Check if index is a string... PASS
* Check index pattern... PASS
* Check for missing values... PASS
* Check for larger start position... PASS.
* Check if zero based... PASS
* Checking sort order... PASS
* Checking for overlapping 'contiguous' regions... PASS
* Processing input (2): b
CONVERT TO BED
* Checking input type... PASS
Input is in bed format
VALIDATE REGIONS
* Check if index is a string... PASS
* Check index pattern... PASS
* Check for missing values... PASS
* Check for larger start position... PASS.
* Check if zero based... PASS
* Checking sort order... PASS
* Checking for overlapping 'contiguous' regions... PASS
bedtools intersect -a /var/folders/rf/qrcw6ncj05z1pc_pq9xzw3540000gn/T//Rtmph6wW6Y/a_c3e316ec6cc5.bed -b /var/folders/rf/qrcw6ncj05z1pc_pq9xzw3540000gn/T//Rtmph6wW6Y/b_c3e379a1c588.bed -wao -f 0.4 -F 0.4summary(as.integer(heart_liver_40[,9]) > 0) Mode FALSE TRUE
logical 18161 4400 # 50% overlap
heart_liver_50 <- bedr(input = list(a = sort_humans_heart_liver_DMRs_bed, b = sort_humans_heart_lung_DMRs_bed), method = "intersect", params = "-wao -f 0.50 -F 0.50") * Processing input (1): a
CONVERT TO BED
* Checking input type... PASS
Input is in bed format
VALIDATE REGIONS
* Check if index is a string... PASS
* Check index pattern... PASS
* Check for missing values... PASS
* Check for larger start position... PASS.
* Check if zero based... PASS
* Checking sort order... PASS
* Checking for overlapping 'contiguous' regions... PASS
* Processing input (2): b
CONVERT TO BED
* Checking input type... PASS
Input is in bed format
VALIDATE REGIONS
* Check if index is a string... PASS
* Check index pattern... PASS
* Check for missing values... PASS
* Check for larger start position... PASS.
* Check if zero based... PASS
* Checking sort order... PASS
* Checking for overlapping 'contiguous' regions... PASS
bedtools intersect -a /var/folders/rf/qrcw6ncj05z1pc_pq9xzw3540000gn/T//Rtmph6wW6Y/a_c3e35af7adf6.bed -b /var/folders/rf/qrcw6ncj05z1pc_pq9xzw3540000gn/T//Rtmph6wW6Y/b_c3e3646a08f0.bed -wao -f 0.50 -F 0.50summary(as.integer(heart_liver_50[,9]) > 0) Mode FALSE TRUE
logical 18703 3858 #60% overlap
heart_liver_60 <- bedr(input = list(a = sort_humans_heart_liver_DMRs_bed, b = sort_humans_heart_lung_DMRs_bed), method = "intersect", params = "-wao -f 0.6 -F 0.6") * Processing input (1): a
CONVERT TO BED
* Checking input type... PASS
Input is in bed format
VALIDATE REGIONS
* Check if index is a string... PASS
* Check index pattern... PASS
* Check for missing values... PASS
* Check for larger start position... PASS.
* Check if zero based... PASS
* Checking sort order... PASS
* Checking for overlapping 'contiguous' regions... PASS
* Processing input (2): b
CONVERT TO BED
* Checking input type... PASS
Input is in bed format
VALIDATE REGIONS
* Check if index is a string... PASS
* Check index pattern... PASS
* Check for missing values... PASS
* Check for larger start position... PASS.
* Check if zero based... PASS
* Checking sort order... PASS
* Checking for overlapping 'contiguous' regions... PASS
bedtools intersect -a /var/folders/rf/qrcw6ncj05z1pc_pq9xzw3540000gn/T//Rtmph6wW6Y/a_c3e3b527156.bed -b /var/folders/rf/qrcw6ncj05z1pc_pq9xzw3540000gn/T//Rtmph6wW6Y/b_c3e34a0fee0e.bed -wao -f 0.6 -F 0.6summary(as.integer(heart_liver_60[,9]) > 0) Mode FALSE TRUE
logical 19286 3275 # 70% overlap
heart_liver_70 <- bedr(input = list(a = sort_humans_heart_liver_DMRs_bed, b = sort_humans_heart_lung_DMRs_bed), method = "intersect", params = "-wao -f 0.70 -F 0.70") * Processing input (1): a
CONVERT TO BED
* Checking input type... PASS
Input is in bed format
VALIDATE REGIONS
* Check if index is a string... PASS
* Check index pattern... PASS
* Check for missing values... PASS
* Check for larger start position... PASS.
* Check if zero based... PASS
* Checking sort order... PASS
* Checking for overlapping 'contiguous' regions... PASS
* Processing input (2): b
CONVERT TO BED
* Checking input type... PASS
Input is in bed format
VALIDATE REGIONS
* Check if index is a string... PASS
* Check index pattern... PASS
* Check for missing values... PASS
* Check for larger start position... PASS.
* Check if zero based... PASS
* Checking sort order... PASS
* Checking for overlapping 'contiguous' regions... PASS
bedtools intersect -a /var/folders/rf/qrcw6ncj05z1pc_pq9xzw3540000gn/T//Rtmph6wW6Y/a_c3e31f6a6b32.bed -b /var/folders/rf/qrcw6ncj05z1pc_pq9xzw3540000gn/T//Rtmph6wW6Y/b_c3e319b55a59.bed -wao -f 0.70 -F 0.70summary(as.integer(heart_liver_70[,9]) > 0) Mode FALSE TRUE
logical 19925 2636 #75% overlap
heart_liver_75 <- bedr(input = list(a = sort_humans_heart_liver_DMRs_bed, b = sort_humans_heart_lung_DMRs_bed), method = "intersect", params = "-wao -f 0.75 -F 0.75") * Processing input (1): a
CONVERT TO BED
* Checking input type... PASS
Input is in bed format
VALIDATE REGIONS
* Check if index is a string... PASS
* Check index pattern... PASS
* Check for missing values... PASS
* Check for larger start position... PASS.
* Check if zero based... PASS
* Checking sort order... PASS
* Checking for overlapping 'contiguous' regions... PASS
* Processing input (2): b
CONVERT TO BED
* Checking input type... PASS
Input is in bed format
VALIDATE REGIONS
* Check if index is a string... PASS
* Check index pattern... PASS
* Check for missing values... PASS
* Check for larger start position... PASS.
* Check if zero based... PASS
* Checking sort order... PASS
* Checking for overlapping 'contiguous' regions... PASS
bedtools intersect -a /var/folders/rf/qrcw6ncj05z1pc_pq9xzw3540000gn/T//Rtmph6wW6Y/a_c3e32c794932.bed -b /var/folders/rf/qrcw6ncj05z1pc_pq9xzw3540000gn/T//Rtmph6wW6Y/b_c3e364556bbc.bed -wao -f 0.75 -F 0.75summary(as.integer(heart_liver_75[,9]) > 0) Mode FALSE TRUE
logical 20256 2305 # 80% overlap
heart_liver_80 <- bedr(input = list(a = sort_humans_heart_liver_DMRs_bed, b = sort_humans_heart_lung_DMRs_bed), method = "intersect", params = "-wao -f 0.80 -F 0.80") * Processing input (1): a
CONVERT TO BED
* Checking input type... PASS
Input is in bed format
VALIDATE REGIONS
* Check if index is a string... PASS
* Check index pattern... PASS
* Check for missing values... PASS
* Check for larger start position... PASS.
* Check if zero based... PASS
* Checking sort order... PASS
* Checking for overlapping 'contiguous' regions... PASS
* Processing input (2): b
CONVERT TO BED
* Checking input type... PASS
Input is in bed format
VALIDATE REGIONS
* Check if index is a string... PASS
* Check index pattern... PASS
* Check for missing values... PASS
* Check for larger start position... PASS.
* Check if zero based... PASS
* Checking sort order... PASS
* Checking for overlapping 'contiguous' regions... PASS
bedtools intersect -a /var/folders/rf/qrcw6ncj05z1pc_pq9xzw3540000gn/T//Rtmph6wW6Y/a_c3e33ffeacc6.bed -b /var/folders/rf/qrcw6ncj05z1pc_pq9xzw3540000gn/T//Rtmph6wW6Y/b_c3e34cda51f0.bed -wao -f 0.80 -F 0.80summary(as.integer(heart_liver_80[,9]) > 0) Mode FALSE TRUE
logical 20597 1964 #90% overlap
heart_liver_90 <- bedr(input = list(a = sort_humans_heart_liver_DMRs_bed, b = sort_humans_heart_lung_DMRs_bed), method = "intersect", params = "-wao -f 0.9 -F 0.9") * Processing input (1): a
CONVERT TO BED
* Checking input type... PASS
Input is in bed format
VALIDATE REGIONS
* Check if index is a string... PASS
* Check index pattern... PASS
* Check for missing values... PASS
* Check for larger start position... PASS.
* Check if zero based... PASS
* Checking sort order... PASS
* Checking for overlapping 'contiguous' regions... PASS
* Processing input (2): b
CONVERT TO BED
* Checking input type... PASS
Input is in bed format
VALIDATE REGIONS
* Check if index is a string... PASS
* Check index pattern... PASS
* Check for missing values... PASS
* Check for larger start position... PASS.
* Check if zero based... PASS
* Checking sort order... PASS
* Checking for overlapping 'contiguous' regions... PASS
bedtools intersect -a /var/folders/rf/qrcw6ncj05z1pc_pq9xzw3540000gn/T//Rtmph6wW6Y/a_c3e320beb9cd.bed -b /var/folders/rf/qrcw6ncj05z1pc_pq9xzw3540000gn/T//Rtmph6wW6Y/b_c3e330ca4304.bed -wao -f 0.9 -F 0.9summary(as.integer(heart_liver_90[,9]) > 0) Mode FALSE TRUE
logical 21223 1338 #100% overlap
heart_liver_100 <- bedr(input = list(a = sort_humans_heart_liver_DMRs_bed, b = sort_humans_heart_lung_DMRs_bed), method = "intersect", params = "-wao -f 1 -F 1") * Processing input (1): a
CONVERT TO BED
* Checking input type... PASS
Input is in bed format
VALIDATE REGIONS
* Check if index is a string... PASS
* Check index pattern... PASS
* Check for missing values... PASS
* Check for larger start position... PASS.
* Check if zero based... PASS
* Checking sort order... PASS
* Checking for overlapping 'contiguous' regions... PASS
* Processing input (2): b
CONVERT TO BED
* Checking input type... PASS
Input is in bed format
VALIDATE REGIONS
* Check if index is a string... PASS
* Check index pattern... PASS
* Check for missing values... PASS
* Check for larger start position... PASS.
* Check if zero based... PASS
* Checking sort order... PASS
* Checking for overlapping 'contiguous' regions... PASS
bedtools intersect -a /var/folders/rf/qrcw6ncj05z1pc_pq9xzw3540000gn/T//Rtmph6wW6Y/a_c3e34d3333b5.bed -b /var/folders/rf/qrcw6ncj05z1pc_pq9xzw3540000gn/T//Rtmph6wW6Y/b_c3e367dc9ea8.bed -wao -f 1 -F 1summary(as.integer(heart_liver_100[,9]) > 0) Mode FALSE TRUE
logical 21587 974 Plot the overlap
# Find the number of tDMRs depending on the bp overlap
summary(as.integer(heart_liver_1[,9]) > 0) Mode FALSE TRUE
logical 16965 5607 summary(as.integer(heart_liver_10[,9]) > 0) Mode FALSE TRUE
logical 17096 5472 summary(as.integer(heart_liver_20[,9]) > 0) Mode FALSE TRUE
logical 17374 5189 summary(as.integer(heart_liver_25[,9]) > 0) Mode FALSE TRUE
logical 17554 5007 summary(as.integer(heart_liver_30[,9]) > 0) Mode FALSE TRUE
logical 17737 4824 summary(as.integer(heart_liver_40[,9]) > 0) Mode FALSE TRUE
logical 18161 4400 summary(as.integer(heart_liver_50[,9]) > 0) Mode FALSE TRUE
logical 18703 3858 summary(as.integer(heart_liver_60[,9]) > 0) Mode FALSE TRUE
logical 19286 3275 summary(as.integer(heart_liver_70[,9]) > 0) Mode FALSE TRUE
logical 19925 2636 summary(as.integer(heart_liver_75[,9]) > 0) Mode FALSE TRUE
logical 20256 2305 summary(as.integer(heart_liver_80[,9]) > 0) Mode FALSE TRUE
logical 20597 1964 summary(as.integer(heart_liver_90[,9]) > 0) Mode FALSE TRUE
logical 21223 1338 summary(as.integer(heart_liver_100[,9]) > 0) Mode FALSE TRUE
logical 21587 974 # Plot the number of tDMRs
conserved_tdmr_heart_liver_heart_lung <- c(5607, 5472, 5189, 5007, 4824, 4400, 3858, 3275, 2636, 2305,1964,1338, 974)
perc_overlap <- c(0.000000001, 0.1, 0.2, 0.25, 0.3, 0.4, 0.5, 0.6, 0.7, 0.75, 0.80, 0.90, 1)
plot(perc_overlap, conserved_tdmr_heart_liver_heart_lung, xlab = "Percentage of overlap (bp)", ylab = "Number of human tDMRs (HvLi and HvLu)", pch = 19)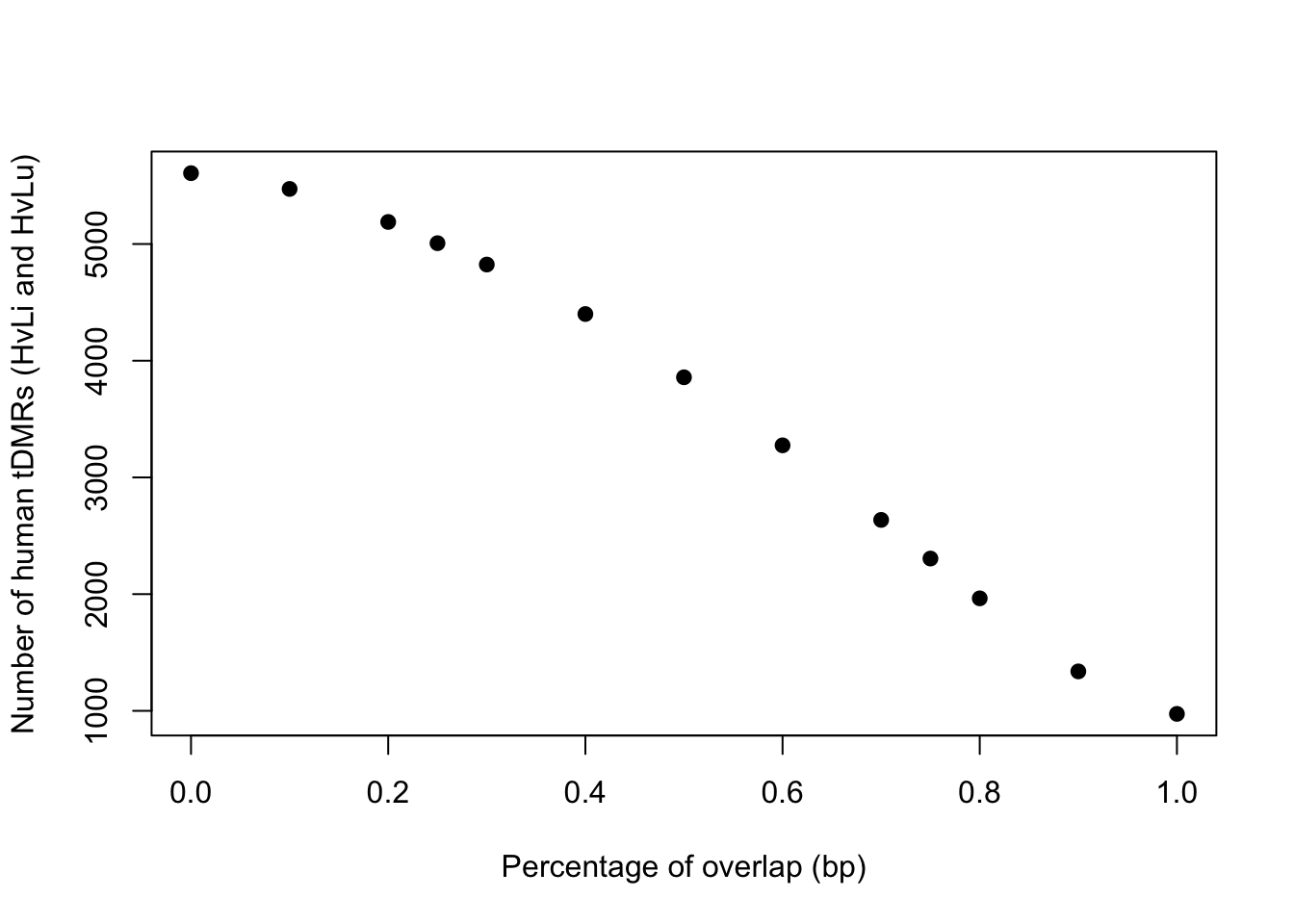
# Investigate the size of the tDMRs depending on the percentage overlap
check_1 <- heart_liver_10[which((as.integer(heart_liver_1[,9]) > 0) == TRUE),]
check_10 <- heart_liver_10[which((as.integer(heart_liver_10[,9]) > 0) == TRUE),]
check_20 <- heart_liver_20[which((as.integer(heart_liver_20[,9]) > 0) == TRUE),]
check_25 <- heart_liver_25[which((as.integer(heart_liver_25[,9]) > 0) == TRUE),]
check_30 <- heart_liver_30[which((as.integer(heart_liver_30[,9]) > 0) == TRUE),]
check_40 <- heart_liver_40[which((as.integer(heart_liver_40[,9]) > 0) == TRUE),]
check_50 <- heart_liver_50[which((as.integer(heart_liver_50[,9]) > 0) == TRUE),]
check_60 <- heart_liver_60[which((as.integer(heart_liver_60[,9]) > 0) == TRUE),]
check_70 <- heart_liver_70[which((as.integer(heart_liver_70[,9]) > 0) == TRUE),]
check_75 <- heart_liver_75[which((as.integer(heart_liver_75[,9]) > 0) == TRUE),]
check_80 <- heart_liver_80[which((as.integer(heart_liver_80[,9]) > 0) == TRUE),]
check_90 <- heart_liver_90[which((as.integer(heart_liver_90[,9]) > 0) == TRUE),]
check_100 <- heart_liver_100[which((as.integer(heart_liver_100[,9]) > 0) == TRUE),]
summary(as.integer(check_1[,9])) Min. 1st Qu. Median Mean 3rd Qu. Max. NA's
0.0 0.0 120.0 280.7 468.8 3167.0 1 summary(as.integer(check_10[,9])) Min. 1st Qu. Median Mean 3rd Qu. Max.
6.0 228.0 419.0 501.7 697.0 3167.0 summary(as.integer(check_20[,9])) Min. 1st Qu. Median Mean 3rd Qu. Max.
6 251 440 522 717 3167 summary(as.integer(check_25[,9])) Min. 1st Qu. Median Mean 3rd Qu. Max.
6.0 265.0 454.0 534.6 727.0 3167.0 summary(as.integer(check_30[,9])) Min. 1st Qu. Median Mean 3rd Qu. Max.
6.0 275.8 470.0 546.2 738.0 3167.0 summary(as.integer(check_40[,9])) Min. 1st Qu. Median Mean 3rd Qu. Max.
6.0 300.0 500.0 570.6 764.0 3167.0 summary(as.integer(check_50[,9])) Min. 1st Qu. Median Mean 3rd Qu. Max.
6.0 323.0 533.0 598.4 806.0 3167.0 summary(as.integer(check_60[,9])) Min. 1st Qu. Median Mean 3rd Qu. Max.
6.0 344.0 574.0 627.7 845.0 3167.0 summary(as.integer(check_70[,9])) Min. 1st Qu. Median Mean 3rd Qu. Max.
6.0 350.0 601.0 649.6 885.5 3167.0 summary(as.integer(check_75[,9])) Min. 1st Qu. Median Mean 3rd Qu. Max.
6.0 346.0 604.0 655.4 904.0 3167.0 summary(as.integer(check_80[,9])) Min. 1st Qu. Median Mean 3rd Qu. Max.
6.0 332.8 599.0 650.3 907.8 3167.0 summary(as.integer(check_90[,9])) Min. 1st Qu. Median Mean 3rd Qu. Max.
6.0 276.5 522.0 601.2 839.0 3167.0 summary(as.integer(check_100[,9])) Min. 1st Qu. Median Mean 3rd Qu. Max.
6.0 223.2 410.5 502.5 707.8 2658.0 # Plot the median tDMR length
conserved_tdmr_heart_liver_heart_lung_bp <- c(120, 419, 440, 454, 470, 500, 533, 574, 601, 604,599,522, 410.5)
perc_overlap <- c(0.000000001, 0.1, 0.2, 0.25, 0.3, 0.4, 0.5, 0.6, 0.7, 0.75, 0.80, 0.90, 1)
plot(perc_overlap, conserved_tdmr_heart_liver_heart_lung_bp, xlab = "Percentage of overlap (bp)", ylab = "Median length of overlap (bp)", pch = 19, main = "Median length of overlap between human tDMRs (HvLi and HvLu)")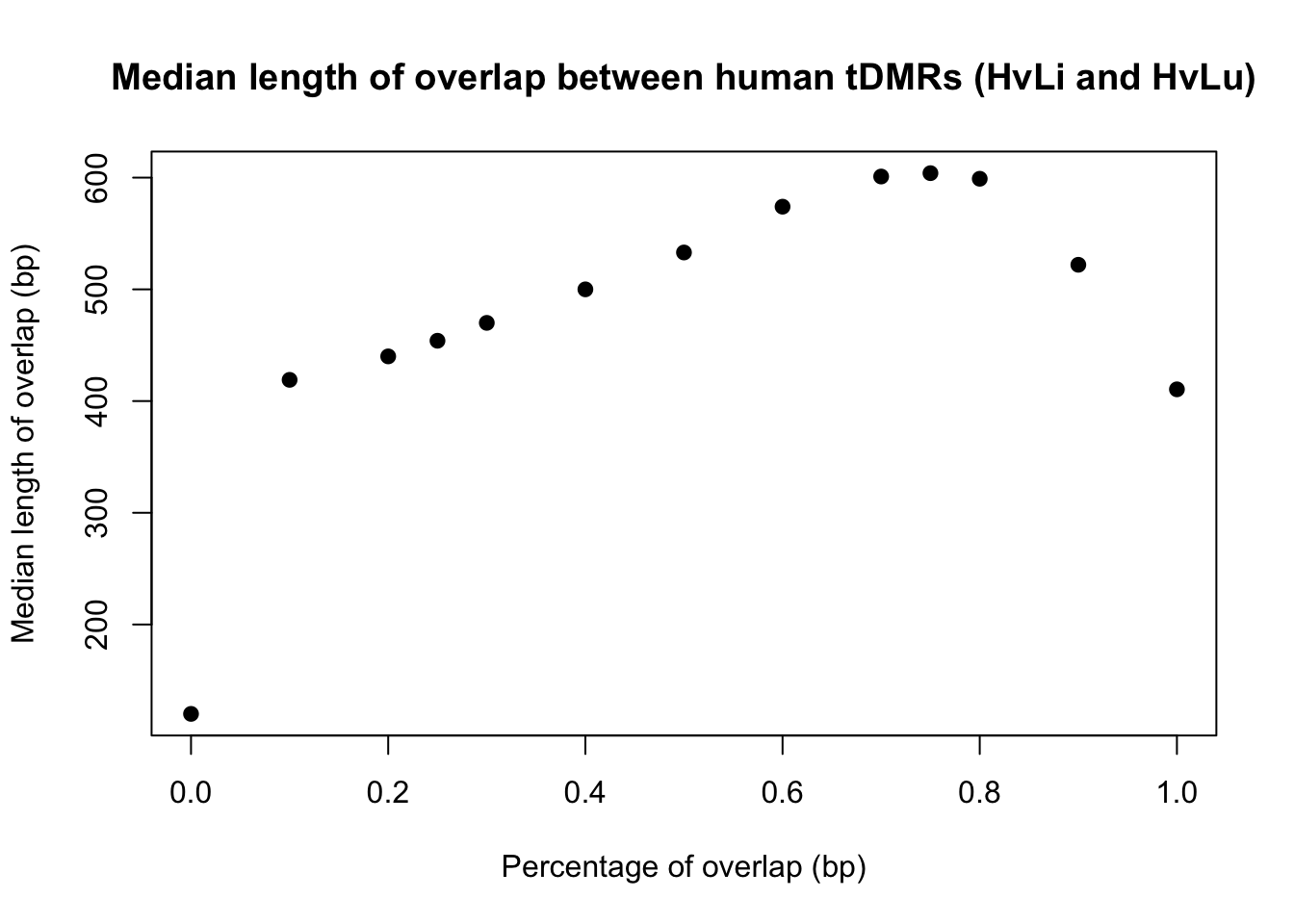
Are the overlap results robust with respect to tissue?
Look at human heart kidney versus human liver lung.
# make bed style format: change name from human to rhesus
humans_heart_liver_DMRs_bed <- humans_heart_kidney_DMRs[,pull_col]
# Get DMRs on the autosomal chromosomes only
humans_heart_liver_DMRs_bed_noX <- humans_heart_liver_DMRs_bed[which(humans_heart_liver_DMRs_bed[,1] != "chrX"),]
humans_heart_liver_DMRs_bed_noX[,1] <- as.character(humans_heart_liver_DMRs_bed_noX[,1])
humans_heart_liver_DMRs_bed_noX[,2] <- as.integer(humans_heart_liver_DMRs_bed_noX[,2])
humans_heart_liver_DMRs_bed_noX[,3] <- as.integer(humans_heart_liver_DMRs_bed_noX[,3])
summary(humans_heart_liver_DMRs_bed_noX[,3] - humans_heart_liver_DMRs_bed_noX[,2]) Min. 1st Qu. Median Mean 3rd Qu. Max.
6.0 261.0 486.0 594.7 831.0 4630.0 sort_humans_heart_liver_DMRs_bed <- bedr.sort.region(humans_heart_liver_DMRs_bed_noX, check.chr = FALSE)SORTING
VALIDATE REGIONS
* Checking input type... PASS
Input is in bed format
* Check if index is a string... PASS
* Check index pattern... PASS
* Check for missing values... PASS
* Check for larger start position... PASS.
* Check if zero based... PASS
* Overlapping regions can cause unexpected results.# make bed style format
humans_heart_lung_DMRs_bed <- humans_liver_lung_DMRs[,pull_col]
humans_heart_lung_DMRs_bed_noX <- humans_heart_lung_DMRs_bed[which(humans_heart_lung_DMRs_bed[,1] != "chrX"),]
humans_heart_lung_DMRs_bed_noX[,1] <- as.character(humans_heart_lung_DMRs_bed_noX[,1])
humans_heart_lung_DMRs_bed_noX[,2] <- as.integer(humans_heart_lung_DMRs_bed_noX[,2])
humans_heart_lung_DMRs_bed_noX[,3] <- as.integer(humans_heart_lung_DMRs_bed_noX[,3])
sort_humans_heart_lung_DMRs_bed <- bedr.sort.region(humans_heart_lung_DMRs_bed_noX, check.chr = FALSE)SORTING
VALIDATE REGIONS
* Checking input type... PASS
Input is in bed format
* Check if index is a string... PASS
* Check index pattern... PASS
* Check for missing values... PASS
* Check for larger start position... PASS.
* Check if zero based... PASS
* Overlapping regions can cause unexpected results.summary(humans_heart_lung_DMRs_bed_noX[,3] - humans_heart_lung_DMRs_bed_noX[,2]) Min. 1st Qu. Median Mean 3rd Qu. Max.
5.0 228.0 427.0 540.7 738.0 4768.0 heart_lung <- bedr(input = list(a = sort_humans_heart_liver_DMRs_bed, b = sort_humans_heart_lung_DMRs_bed), method = "intersect", params = "-wao -f 0.5 -F 0.5", check.chr = FALSE) * Processing input (1): a
CONVERT TO BED
* Checking input type... PASS
Input is in bed format
VALIDATE REGIONS
* Check if index is a string... PASS
* Check index pattern... PASS
* Check for missing values... PASS
* Check for larger start position... PASS.
* Check if zero based... PASS
* Checking sort order... PASS
* Checking for overlapping 'contiguous' regions... PASS
* Processing input (2): b
CONVERT TO BED
* Checking input type... PASS
Input is in bed format
VALIDATE REGIONS
* Check if index is a string... PASS
* Check index pattern... PASS
* Check for missing values... PASS
* Check for larger start position... PASS.
* Check if zero based... PASS
* Checking sort order... PASS
* Checking for overlapping 'contiguous' regions... PASS
bedtools intersect -a /var/folders/rf/qrcw6ncj05z1pc_pq9xzw3540000gn/T//Rtmph6wW6Y/a_c3e31051e7b6.bed -b /var/folders/rf/qrcw6ncj05z1pc_pq9xzw3540000gn/T//Rtmph6wW6Y/b_c3e34c8a8a31.bed -wao -f 0.5 -F 0.5summary(heart_lung[,3] - heart_lung[,2]) Min. 1st Qu. Median Mean 3rd Qu. Max.
6.0 261.0 486.0 594.7 831.0 4630.0 summary(as.integer(heart_lung[,9])) Min. 1st Qu. Median Mean 3rd Qu. Max.
0.00 0.00 0.00 20.34 0.00 2071.00 summary(as.integer(heart_lung[,9]) > 0) Mode FALSE TRUE
logical 29136 1155 lung_heart <- bedr(input = list(a = sort_humans_heart_lung_DMRs_bed, b = sort_humans_heart_liver_DMRs_bed), method = "intersect", params = "-wao -f 0.5 -F 0.5", check.chr = FALSE) * Processing input (1): a
CONVERT TO BED
* Checking input type... PASS
Input is in bed format
VALIDATE REGIONS
* Check if index is a string... PASS
* Check index pattern... PASS
* Check for missing values... PASS
* Check for larger start position... PASS.
* Check if zero based... PASS
* Checking sort order... PASS
* Checking for overlapping 'contiguous' regions... PASS
* Processing input (2): b
CONVERT TO BED
* Checking input type... PASS
Input is in bed format
VALIDATE REGIONS
* Check if index is a string... PASS
* Check index pattern... PASS
* Check for missing values... PASS
* Check for larger start position... PASS.
* Check if zero based... PASS
* Checking sort order... PASS
* Checking for overlapping 'contiguous' regions... PASS
bedtools intersect -a /var/folders/rf/qrcw6ncj05z1pc_pq9xzw3540000gn/T//Rtmph6wW6Y/a_c3e35924cc9b.bed -b /var/folders/rf/qrcw6ncj05z1pc_pq9xzw3540000gn/T//Rtmph6wW6Y/b_c3e36fd217d4.bed -wao -f 0.5 -F 0.5summary(as.integer(lung_heart[,9])) Min. 1st Qu. Median Mean 3rd Qu. Max.
0.00 0.00 0.00 47.99 0.00 2071.00 summary(as.integer(lung_heart[,9]) > 0) Mode FALSE TRUE
logical 11687 1155 # Make a Venn diagram based on the results
summary(as.integer(heart_lung[,9]) > 0) Mode FALSE TRUE
logical 29136 1155 summary(as.integer(lung_heart[,9]) > 0) Mode FALSE TRUE
logical 11687 1155 grid.newpage()
draw.pairwise.venn(area1 = (29136+1155), area2 = (11687+1155), cross.area = 1155, category = c("Heart v. Liver", "Heart v. Lung"))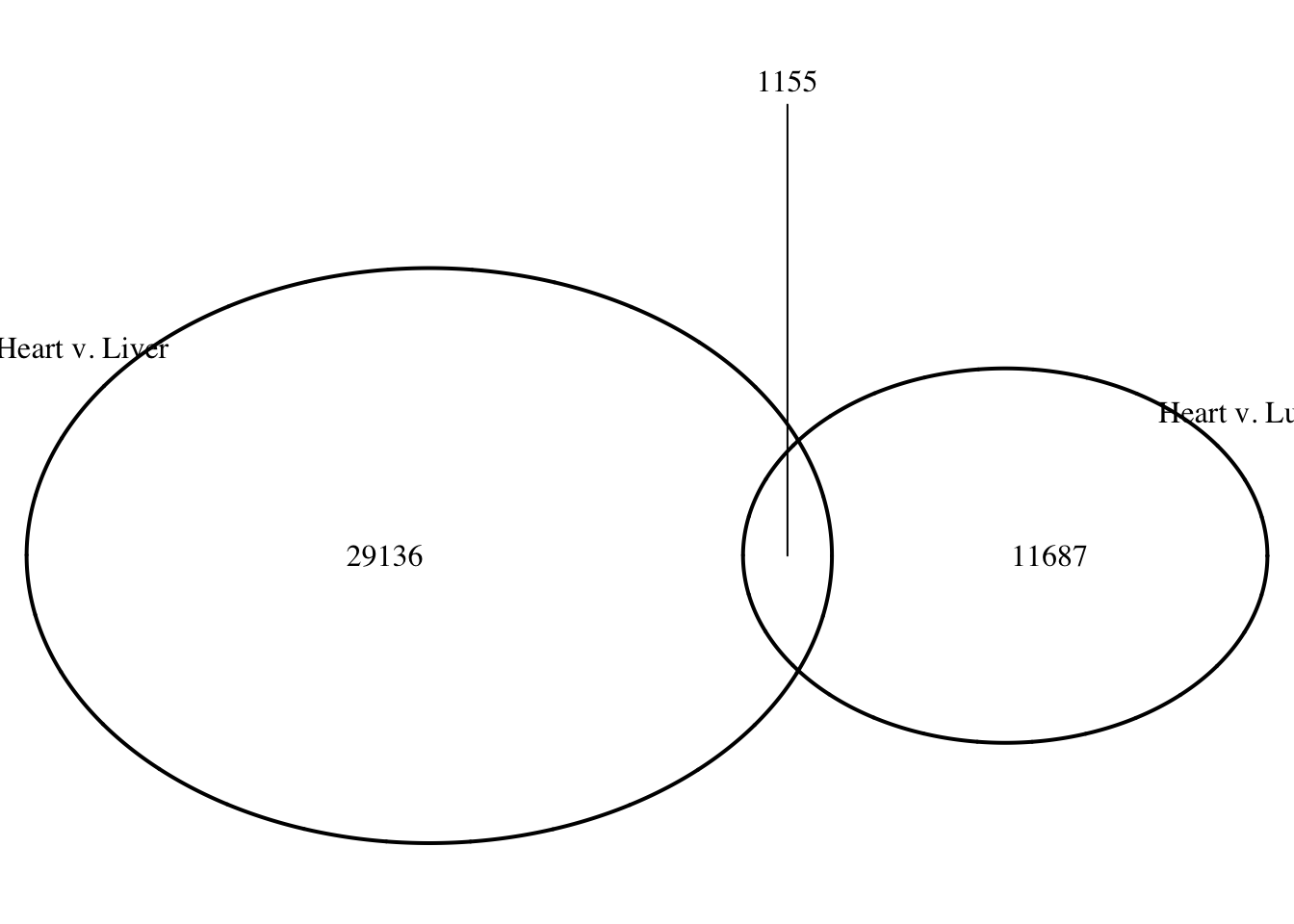
(polygon[GRID.polygon.10], polygon[GRID.polygon.11], polygon[GRID.polygon.12], polygon[GRID.polygon.13], text[GRID.text.14], text[GRID.text.15], text[GRID.text.16], lines[GRID.lines.17], text[GRID.text.18], text[GRID.text.19]) # 1 bp overlap
heart_liver_1 <- bedr(input = list(a = sort_humans_heart_liver_DMRs_bed, b = sort_humans_heart_lung_DMRs_bed), method = "intersect", params = "-wao -f 0.00000001 -F 0.00000001") * Processing input (1): a
CONVERT TO BED
* Checking input type... PASS
Input is in bed format
VALIDATE REGIONS
* Check if index is a string... PASS
* Check index pattern... PASS
* Check for missing values... PASS
* Check for larger start position... PASS.
* Check if zero based... PASS
* Checking sort order... PASS
* Checking for overlapping 'contiguous' regions... PASS
* Processing input (2): b
CONVERT TO BED
* Checking input type... PASS
Input is in bed format
VALIDATE REGIONS
* Check if index is a string... PASS
* Check index pattern... PASS
* Check for missing values... PASS
* Check for larger start position... PASS.
* Check if zero based... PASS
* Checking sort order... PASS
* Checking for overlapping 'contiguous' regions... PASS
bedtools intersect -a /var/folders/rf/qrcw6ncj05z1pc_pq9xzw3540000gn/T//Rtmph6wW6Y/a_c3e345c3354a.bed -b /var/folders/rf/qrcw6ncj05z1pc_pq9xzw3540000gn/T//Rtmph6wW6Y/b_c3e356e266fc.bed -wao -f 0.00000001 -F 0.00000001# 10% overlap
heart_liver_10 <- bedr(input = list(a = sort_humans_heart_liver_DMRs_bed, b = sort_humans_heart_lung_DMRs_bed), method = "intersect", params = "-wao -f 0.10 -F 0.10", check.chr = FALSE) * Processing input (1): a
CONVERT TO BED
* Checking input type... PASS
Input is in bed format
VALIDATE REGIONS
* Check if index is a string... PASS
* Check index pattern... PASS
* Check for missing values... PASS
* Check for larger start position... PASS.
* Check if zero based... PASS
* Checking sort order... PASS
* Checking for overlapping 'contiguous' regions... PASS
* Processing input (2): b
CONVERT TO BED
* Checking input type... PASS
Input is in bed format
VALIDATE REGIONS
* Check if index is a string... PASS
* Check index pattern... PASS
* Check for missing values... PASS
* Check for larger start position... PASS.
* Check if zero based... PASS
* Checking sort order... PASS
* Checking for overlapping 'contiguous' regions... PASS
bedtools intersect -a /var/folders/rf/qrcw6ncj05z1pc_pq9xzw3540000gn/T//Rtmph6wW6Y/a_c3e322e515dc.bed -b /var/folders/rf/qrcw6ncj05z1pc_pq9xzw3540000gn/T//Rtmph6wW6Y/b_c3e339813a85.bed -wao -f 0.10 -F 0.10summary(as.integer(heart_liver_10[,9]) > 0) Mode FALSE TRUE
logical 28021 2275 # 20% overlap
heart_liver_20 <- bedr(input = list(a = sort_humans_heart_liver_DMRs_bed, b = sort_humans_heart_lung_DMRs_bed), method = "intersect", params = "-wao -f 0.20 -F 0.20", check.chr = FALSE) * Processing input (1): a
CONVERT TO BED
* Checking input type... PASS
Input is in bed format
VALIDATE REGIONS
* Check if index is a string... PASS
* Check index pattern... PASS
* Check for missing values... PASS
* Check for larger start position... PASS.
* Check if zero based... PASS
* Checking sort order... PASS
* Checking for overlapping 'contiguous' regions... PASS
* Processing input (2): b
CONVERT TO BED
* Checking input type... PASS
Input is in bed format
VALIDATE REGIONS
* Check if index is a string... PASS
* Check index pattern... PASS
* Check for missing values... PASS
* Check for larger start position... PASS.
* Check if zero based... PASS
* Checking sort order... PASS
* Checking for overlapping 'contiguous' regions... PASS
bedtools intersect -a /var/folders/rf/qrcw6ncj05z1pc_pq9xzw3540000gn/T//Rtmph6wW6Y/a_c3e372626709.bed -b /var/folders/rf/qrcw6ncj05z1pc_pq9xzw3540000gn/T//Rtmph6wW6Y/b_c3e37080530d.bed -wao -f 0.20 -F 0.20summary(as.integer(heart_liver_20[,9]) > 0) Mode FALSE TRUE
logical 28295 1998 # 25% overlap
heart_liver_25 <- bedr(input = list(a = sort_humans_heart_liver_DMRs_bed, b = sort_humans_heart_lung_DMRs_bed), method = "intersect", params = "-wao -f 0.25 -F 0.25", check.chr = FALSE) * Processing input (1): a
CONVERT TO BED
* Checking input type... PASS
Input is in bed format
VALIDATE REGIONS
* Check if index is a string... PASS
* Check index pattern... PASS
* Check for missing values... PASS
* Check for larger start position... PASS.
* Check if zero based... PASS
* Checking sort order... PASS
* Checking for overlapping 'contiguous' regions... PASS
* Processing input (2): b
CONVERT TO BED
* Checking input type... PASS
Input is in bed format
VALIDATE REGIONS
* Check if index is a string... PASS
* Check index pattern... PASS
* Check for missing values... PASS
* Check for larger start position... PASS.
* Check if zero based... PASS
* Checking sort order... PASS
* Checking for overlapping 'contiguous' regions... PASS
bedtools intersect -a /var/folders/rf/qrcw6ncj05z1pc_pq9xzw3540000gn/T//Rtmph6wW6Y/a_c3e327dfcfb1.bed -b /var/folders/rf/qrcw6ncj05z1pc_pq9xzw3540000gn/T//Rtmph6wW6Y/b_c3e321ea0ca1.bed -wao -f 0.25 -F 0.25summary(as.integer(heart_liver_25[,9]) > 0) Mode FALSE TRUE
logical 28422 1870 # 30% overlap
heart_liver_30 <- bedr(input = list(a = sort_humans_heart_liver_DMRs_bed, b = sort_humans_heart_lung_DMRs_bed), method = "intersect", params = "-wao -f 0.30 -F 0.30", check.chr = FALSE) * Processing input (1): a
CONVERT TO BED
* Checking input type... PASS
Input is in bed format
VALIDATE REGIONS
* Check if index is a string... PASS
* Check index pattern... PASS
* Check for missing values... PASS
* Check for larger start position... PASS.
* Check if zero based... PASS
* Checking sort order... PASS
* Checking for overlapping 'contiguous' regions... PASS
* Processing input (2): b
CONVERT TO BED
* Checking input type... PASS
Input is in bed format
VALIDATE REGIONS
* Check if index is a string... PASS
* Check index pattern... PASS
* Check for missing values... PASS
* Check for larger start position... PASS.
* Check if zero based... PASS
* Checking sort order... PASS
* Checking for overlapping 'contiguous' regions... PASS
bedtools intersect -a /var/folders/rf/qrcw6ncj05z1pc_pq9xzw3540000gn/T//Rtmph6wW6Y/a_c3e398145c.bed -b /var/folders/rf/qrcw6ncj05z1pc_pq9xzw3540000gn/T//Rtmph6wW6Y/b_c3e33ccdfb7b.bed -wao -f 0.30 -F 0.30summary(as.integer(heart_liver_30[,9]) > 0) Mode FALSE TRUE
logical 28552 1739 #40% overlap
heart_liver_40 <- bedr(input = list(a = sort_humans_heart_liver_DMRs_bed, b = sort_humans_heart_lung_DMRs_bed), method = "intersect", params = "-wao -f 0.4 -F 0.4", check.chr = FALSE) * Processing input (1): a
CONVERT TO BED
* Checking input type... PASS
Input is in bed format
VALIDATE REGIONS
* Check if index is a string... PASS
* Check index pattern... PASS
* Check for missing values... PASS
* Check for larger start position... PASS.
* Check if zero based... PASS
* Checking sort order... PASS
* Checking for overlapping 'contiguous' regions... PASS
* Processing input (2): b
CONVERT TO BED
* Checking input type... PASS
Input is in bed format
VALIDATE REGIONS
* Check if index is a string... PASS
* Check index pattern... PASS
* Check for missing values... PASS
* Check for larger start position... PASS.
* Check if zero based... PASS
* Checking sort order... PASS
* Checking for overlapping 'contiguous' regions... PASS
bedtools intersect -a /var/folders/rf/qrcw6ncj05z1pc_pq9xzw3540000gn/T//Rtmph6wW6Y/a_c3e326f8c79.bed -b /var/folders/rf/qrcw6ncj05z1pc_pq9xzw3540000gn/T//Rtmph6wW6Y/b_c3e34f4586b0.bed -wao -f 0.4 -F 0.4summary(as.integer(heart_liver_40[,9]) > 0) Mode FALSE TRUE
logical 28840 1451 # 50% overlap
heart_liver_50 <- bedr(input = list(a = sort_humans_heart_liver_DMRs_bed, b = sort_humans_heart_lung_DMRs_bed), method = "intersect", params = "-wao -f 0.50 -F 0.50", check.chr = FALSE) * Processing input (1): a
CONVERT TO BED
* Checking input type... PASS
Input is in bed format
VALIDATE REGIONS
* Check if index is a string... PASS
* Check index pattern... PASS
* Check for missing values... PASS
* Check for larger start position... PASS.
* Check if zero based... PASS
* Checking sort order... PASS
* Checking for overlapping 'contiguous' regions... PASS
* Processing input (2): b
CONVERT TO BED
* Checking input type... PASS
Input is in bed format
VALIDATE REGIONS
* Check if index is a string... PASS
* Check index pattern... PASS
* Check for missing values... PASS
* Check for larger start position... PASS.
* Check if zero based... PASS
* Checking sort order... PASS
* Checking for overlapping 'contiguous' regions... PASS
bedtools intersect -a /var/folders/rf/qrcw6ncj05z1pc_pq9xzw3540000gn/T//Rtmph6wW6Y/a_c3e36c2350a6.bed -b /var/folders/rf/qrcw6ncj05z1pc_pq9xzw3540000gn/T//Rtmph6wW6Y/b_c3e31c7e8db7.bed -wao -f 0.50 -F 0.50summary(as.integer(heart_liver_50[,9]) > 0) Mode FALSE TRUE
logical 29136 1155 #60% overlap
heart_liver_60 <- bedr(input = list(a = sort_humans_heart_liver_DMRs_bed, b = sort_humans_heart_lung_DMRs_bed), method = "intersect", params = "-wao -f 0.6 -F 0.6", check.chr = FALSE) * Processing input (1): a
CONVERT TO BED
* Checking input type... PASS
Input is in bed format
VALIDATE REGIONS
* Check if index is a string... PASS
* Check index pattern... PASS
* Check for missing values... PASS
* Check for larger start position... PASS.
* Check if zero based... PASS
* Checking sort order... PASS
* Checking for overlapping 'contiguous' regions... PASS
* Processing input (2): b
CONVERT TO BED
* Checking input type... PASS
Input is in bed format
VALIDATE REGIONS
* Check if index is a string... PASS
* Check index pattern... PASS
* Check for missing values... PASS
* Check for larger start position... PASS.
* Check if zero based... PASS
* Checking sort order... PASS
* Checking for overlapping 'contiguous' regions... PASS
bedtools intersect -a /var/folders/rf/qrcw6ncj05z1pc_pq9xzw3540000gn/T//Rtmph6wW6Y/a_c3e331d5e1d1.bed -b /var/folders/rf/qrcw6ncj05z1pc_pq9xzw3540000gn/T//Rtmph6wW6Y/b_c3e36413c29a.bed -wao -f 0.6 -F 0.6summary(as.integer(heart_liver_60[,9]) > 0) Mode FALSE TRUE
logical 29425 866 # 70% overlap
heart_liver_70 <- bedr(input = list(a = sort_humans_heart_liver_DMRs_bed, b = sort_humans_heart_lung_DMRs_bed), method = "intersect", params = "-wao -f 0.70 -F 0.70", check.chr = FALSE) * Processing input (1): a
CONVERT TO BED
* Checking input type... PASS
Input is in bed format
VALIDATE REGIONS
* Check if index is a string... PASS
* Check index pattern... PASS
* Check for missing values... PASS
* Check for larger start position... PASS.
* Check if zero based... PASS
* Checking sort order... PASS
* Checking for overlapping 'contiguous' regions... PASS
* Processing input (2): b
CONVERT TO BED
* Checking input type... PASS
Input is in bed format
VALIDATE REGIONS
* Check if index is a string... PASS
* Check index pattern... PASS
* Check for missing values... PASS
* Check for larger start position... PASS.
* Check if zero based... PASS
* Checking sort order... PASS
* Checking for overlapping 'contiguous' regions... PASS
bedtools intersect -a /var/folders/rf/qrcw6ncj05z1pc_pq9xzw3540000gn/T//Rtmph6wW6Y/a_c3e34333d9e2.bed -b /var/folders/rf/qrcw6ncj05z1pc_pq9xzw3540000gn/T//Rtmph6wW6Y/b_c3e3acdadd.bed -wao -f 0.70 -F 0.70summary(as.integer(heart_liver_70[,9]) > 0) Mode FALSE TRUE
logical 29659 632 #75% overlap
heart_liver_75 <- bedr(input = list(a = sort_humans_heart_liver_DMRs_bed, b = sort_humans_heart_lung_DMRs_bed), method = "intersect", params = "-wao -f 0.75 -F 0.75", check.chr = FALSE) * Processing input (1): a
CONVERT TO BED
* Checking input type... PASS
Input is in bed format
VALIDATE REGIONS
* Check if index is a string... PASS
* Check index pattern... PASS
* Check for missing values... PASS
* Check for larger start position... PASS.
* Check if zero based... PASS
* Checking sort order... PASS
* Checking for overlapping 'contiguous' regions... PASS
* Processing input (2): b
CONVERT TO BED
* Checking input type... PASS
Input is in bed format
VALIDATE REGIONS
* Check if index is a string... PASS
* Check index pattern... PASS
* Check for missing values... PASS
* Check for larger start position... PASS.
* Check if zero based... PASS
* Checking sort order... PASS
* Checking for overlapping 'contiguous' regions... PASS
bedtools intersect -a /var/folders/rf/qrcw6ncj05z1pc_pq9xzw3540000gn/T//Rtmph6wW6Y/a_c3e3a0f9d85.bed -b /var/folders/rf/qrcw6ncj05z1pc_pq9xzw3540000gn/T//Rtmph6wW6Y/b_c3e36bf07ca.bed -wao -f 0.75 -F 0.75summary(as.integer(heart_liver_75[,9]) > 0) Mode FALSE TRUE
logical 29784 507 # 80% overlap
heart_liver_80 <- bedr(input = list(a = sort_humans_heart_liver_DMRs_bed, b = sort_humans_heart_lung_DMRs_bed), method = "intersect", params = "-wao -f 0.80 -F 0.80", check.chr = FALSE) * Processing input (1): a
CONVERT TO BED
* Checking input type... PASS
Input is in bed format
VALIDATE REGIONS
* Check if index is a string... PASS
* Check index pattern... PASS
* Check for missing values... PASS
* Check for larger start position... PASS.
* Check if zero based... PASS
* Checking sort order... PASS
* Checking for overlapping 'contiguous' regions... PASS
* Processing input (2): b
CONVERT TO BED
* Checking input type... PASS
Input is in bed format
VALIDATE REGIONS
* Check if index is a string... PASS
* Check index pattern... PASS
* Check for missing values... PASS
* Check for larger start position... PASS.
* Check if zero based... PASS
* Checking sort order... PASS
* Checking for overlapping 'contiguous' regions... PASS
bedtools intersect -a /var/folders/rf/qrcw6ncj05z1pc_pq9xzw3540000gn/T//Rtmph6wW6Y/a_c3e367a8464a.bed -b /var/folders/rf/qrcw6ncj05z1pc_pq9xzw3540000gn/T//Rtmph6wW6Y/b_c3e33a164000.bed -wao -f 0.80 -F 0.80summary(as.integer(heart_liver_80[,9]) > 0) Mode FALSE TRUE
logical 29865 426 #90% overlap
heart_liver_90 <- bedr(input = list(a = sort_humans_heart_liver_DMRs_bed, b = sort_humans_heart_lung_DMRs_bed), method = "intersect", params = "-wao -f 0.9 -F 0.9", check.chr = FALSE) * Processing input (1): a
CONVERT TO BED
* Checking input type... PASS
Input is in bed format
VALIDATE REGIONS
* Check if index is a string... PASS
* Check index pattern... PASS
* Check for missing values... PASS
* Check for larger start position... PASS.
* Check if zero based... PASS
* Checking sort order... PASS
* Checking for overlapping 'contiguous' regions... PASS
* Processing input (2): b
CONVERT TO BED
* Checking input type... PASS
Input is in bed format
VALIDATE REGIONS
* Check if index is a string... PASS
* Check index pattern... PASS
* Check for missing values... PASS
* Check for larger start position... PASS.
* Check if zero based... PASS
* Checking sort order... PASS
* Checking for overlapping 'contiguous' regions... PASS
bedtools intersect -a /var/folders/rf/qrcw6ncj05z1pc_pq9xzw3540000gn/T//Rtmph6wW6Y/a_c3e31bc04a40.bed -b /var/folders/rf/qrcw6ncj05z1pc_pq9xzw3540000gn/T//Rtmph6wW6Y/b_c3e36099fa53.bed -wao -f 0.9 -F 0.9summary(as.integer(heart_liver_90[,9]) > 0) Mode FALSE TRUE
logical 29997 294 #100% overlap
heart_liver_100 <- bedr(input = list(a = sort_humans_heart_liver_DMRs_bed, b = sort_humans_heart_lung_DMRs_bed), method = "intersect", params = "-wao -f 1 -F 1", check.chr = FALSE) * Processing input (1): a
CONVERT TO BED
* Checking input type... PASS
Input is in bed format
VALIDATE REGIONS
* Check if index is a string... PASS
* Check index pattern... PASS
* Check for missing values... PASS
* Check for larger start position... PASS.
* Check if zero based... PASS
* Checking sort order... PASS
* Checking for overlapping 'contiguous' regions... PASS
* Processing input (2): b
CONVERT TO BED
* Checking input type... PASS
Input is in bed format
VALIDATE REGIONS
* Check if index is a string... PASS
* Check index pattern... PASS
* Check for missing values... PASS
* Check for larger start position... PASS.
* Check if zero based... PASS
* Checking sort order... PASS
* Checking for overlapping 'contiguous' regions... PASS
bedtools intersect -a /var/folders/rf/qrcw6ncj05z1pc_pq9xzw3540000gn/T//Rtmph6wW6Y/a_c3e331080eb0.bed -b /var/folders/rf/qrcw6ncj05z1pc_pq9xzw3540000gn/T//Rtmph6wW6Y/b_c3e31e8f19ef.bed -wao -f 1 -F 1summary(as.integer(heart_liver_100[,9]) > 0) Mode FALSE TRUE
logical 30057 234 # Find the number of tDMRs depending on the bp overlap
summary(as.integer(heart_liver_1[,9]) > 0) Mode FALSE TRUE
logical 27864 2440 summary(as.integer(heart_liver_10[,9]) > 0) Mode FALSE TRUE
logical 28021 2275 summary(as.integer(heart_liver_20[,9]) > 0) Mode FALSE TRUE
logical 28295 1998 summary(as.integer(heart_liver_25[,9]) > 0) Mode FALSE TRUE
logical 28422 1870 summary(as.integer(heart_liver_30[,9]) > 0) Mode FALSE TRUE
logical 28552 1739 summary(as.integer(heart_liver_40[,9]) > 0) Mode FALSE TRUE
logical 28840 1451 summary(as.integer(heart_liver_50[,9]) > 0) Mode FALSE TRUE
logical 29136 1155 summary(as.integer(heart_liver_60[,9]) > 0) Mode FALSE TRUE
logical 29425 866 summary(as.integer(heart_liver_70[,9]) > 0) Mode FALSE TRUE
logical 29659 632 summary(as.integer(heart_liver_75[,9]) > 0) Mode FALSE TRUE
logical 29784 507 summary(as.integer(heart_liver_80[,9]) > 0) Mode FALSE TRUE
logical 29865 426 summary(as.integer(heart_liver_90[,9]) > 0) Mode FALSE TRUE
logical 29997 294 summary(as.integer(heart_liver_100[,9]) > 0) Mode FALSE TRUE
logical 30057 234 # Plot the number of tDMRs
conserved_tdmr_heart_liver_heart_lung <- c(2440, 2275, 1998, 1870, 1739, 1451, 1155, 866, 632, 507,426,294, 234)
perc_overlap <- c(0.000000001, 0.1, 0.2, 0.25, 0.3, 0.4, 0.5, 0.6, 0.7, 0.75, 0.80, 0.90, 1)
plot(perc_overlap, conserved_tdmr_heart_liver_heart_lung, xlab = "Percentage of overlap (bp)", ylab = "Number of overlapping chimp tDMRs", pch = 19, main = "Overlapping Human tDMRs (HvK and LivLu)")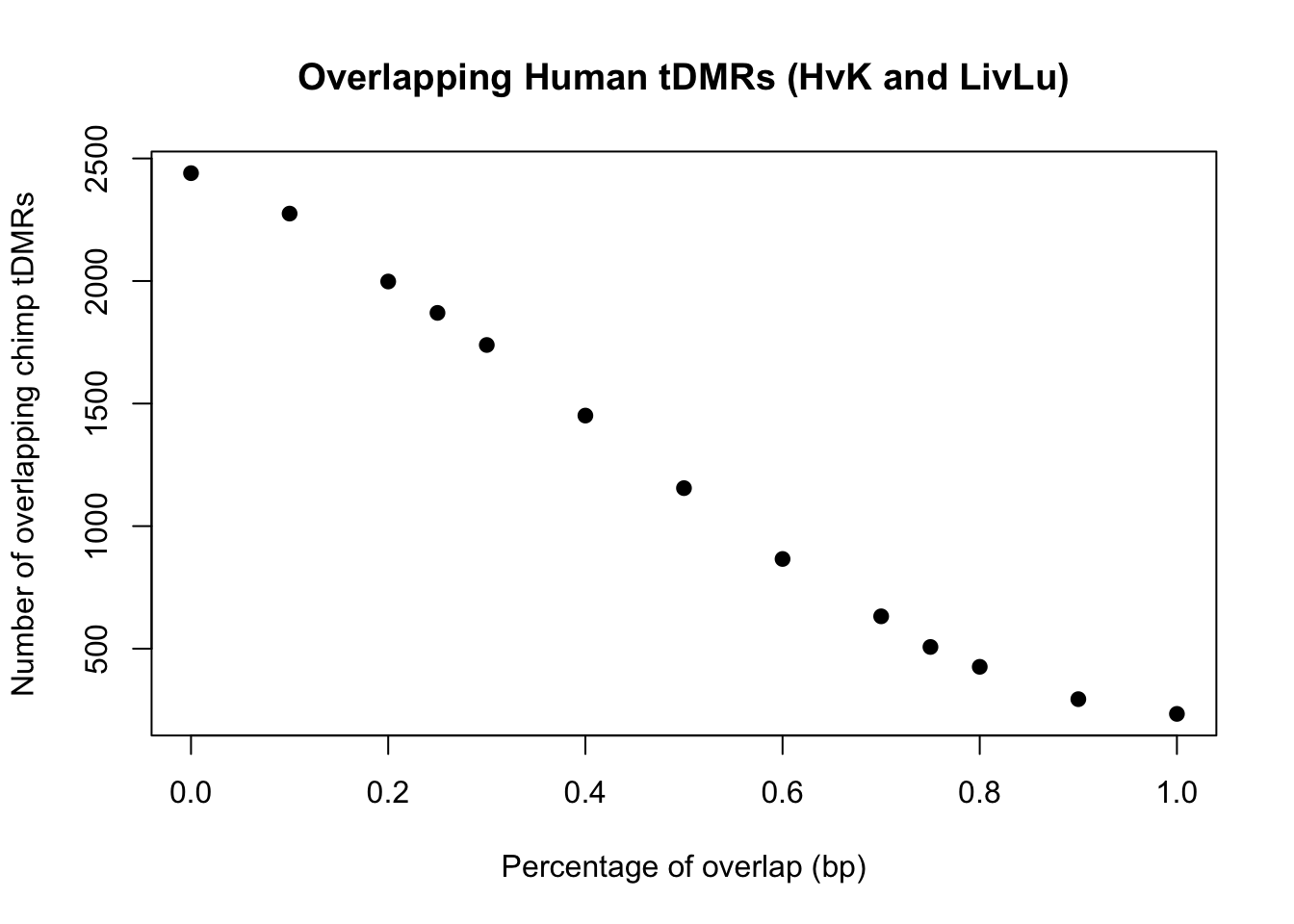
# Investigate the size of the tDMRs depending on the percentage overlap
check_1 <- heart_liver_1[which((as.integer(heart_liver_1[,9]) > 0) == TRUE),]
check_10 <- heart_liver_10[which((as.integer(heart_liver_10[,9]) > 0) == TRUE),]
check_20 <- heart_liver_20[which((as.integer(heart_liver_20[,9]) > 0) == TRUE),]
check_25 <- heart_liver_25[which((as.integer(heart_liver_25[,9]) > 0) == TRUE),]
check_30 <- heart_liver_30[which((as.integer(heart_liver_30[,9]) > 0) == TRUE),]
check_40 <- heart_liver_40[which((as.integer(heart_liver_40[,9]) > 0) == TRUE),]
check_50 <- heart_liver_50[which((as.integer(heart_liver_50[,9]) > 0) == TRUE),]
check_60 <- heart_liver_60[which((as.integer(heart_liver_60[,9]) > 0) == TRUE),]
check_70 <- heart_liver_70[which((as.integer(heart_liver_70[,9]) > 0) == TRUE),]
check_75 <- heart_liver_75[which((as.integer(heart_liver_75[,9]) > 0) == TRUE),]
check_80 <- heart_liver_80[which((as.integer(heart_liver_80[,9]) > 0) == TRUE),]
check_90 <- heart_liver_90[which((as.integer(heart_liver_90[,9]) > 0) == TRUE),]
check_100 <- heart_liver_100[which((as.integer(heart_liver_100[,9]) > 0) == TRUE),]
summary(as.integer(check_1[,9])) Min. 1st Qu. Median Mean 3rd Qu. Max.
2.0 151.8 301.0 378.3 522.0 2071.0 summary(as.integer(check_10[,9])) Min. 1st Qu. Median Mean 3rd Qu. Max.
8.0 173.0 321.0 401.2 545.0 2071.0 summary(as.integer(check_20[,9])) Min. 1st Qu. Median Mean 3rd Qu. Max.
8.0 205.2 357.0 435.5 591.8 2071.0 summary(as.integer(check_25[,9])) Min. 1st Qu. Median Mean 3rd Qu. Max.
8.0 221.0 376.0 450.9 618.8 2071.0 summary(as.integer(check_30[,9])) Min. 1st Qu. Median Mean 3rd Qu. Max.
12.0 233.0 394.0 466.2 635.0 2071.0 summary(as.integer(check_40[,9])) Min. 1st Qu. Median Mean 3rd Qu. Max.
12.0 256.5 429.0 500.2 685.5 2071.0 summary(as.integer(check_50[,9])) Min. 1st Qu. Median Mean 3rd Qu. Max.
12.0 274.0 467.0 533.6 724.0 2071.0 summary(as.integer(check_60[,9])) Min. 1st Qu. Median Mean 3rd Qu. Max.
12.0 274.2 498.5 555.9 760.0 2071.0 summary(as.integer(check_70[,9])) Min. 1st Qu. Median Mean 3rd Qu. Max.
12.0 274.0 503.5 568.9 781.2 2071.0 summary(as.integer(check_75[,9])) Min. 1st Qu. Median Mean 3rd Qu. Max.
12.0 265.0 514.0 577.6 806.0 2071.0 summary(as.integer(check_80[,9])) Min. 1st Qu. Median Mean 3rd Qu. Max.
12.0 240.0 475.5 555.9 801.2 2071.0 summary(as.integer(check_90[,9])) Min. 1st Qu. Median Mean 3rd Qu. Max.
12.0 189.2 390.5 499.5 733.2 2071.0 summary(as.integer(check_100[,9])) Min. 1st Qu. Median Mean 3rd Qu. Max.
12.0 150.0 340.0 434.6 631.8 1866.0 # Plot the median tDMR length
conserved_tdmr_heart_liver_heart_lung_bp <- c(301, 321, 357, 376, 394, 429, 467, 498.5, 503.5, 514, 475.5, 390.5, 340)
perc_overlap <- c(0.000000001, 0.1, 0.2, 0.25, 0.3, 0.4, 0.5, 0.6, 0.7, 0.75, 0.80, 0.90, 1)
plot(perc_overlap, conserved_tdmr_heart_liver_heart_lung_bp, xlab = "Percentage of overlap (bp)", ylab = "Median length of overlap (bp)", pch = 19, main = "Median length of overlap between human tDMRs (HvK and LivLu)")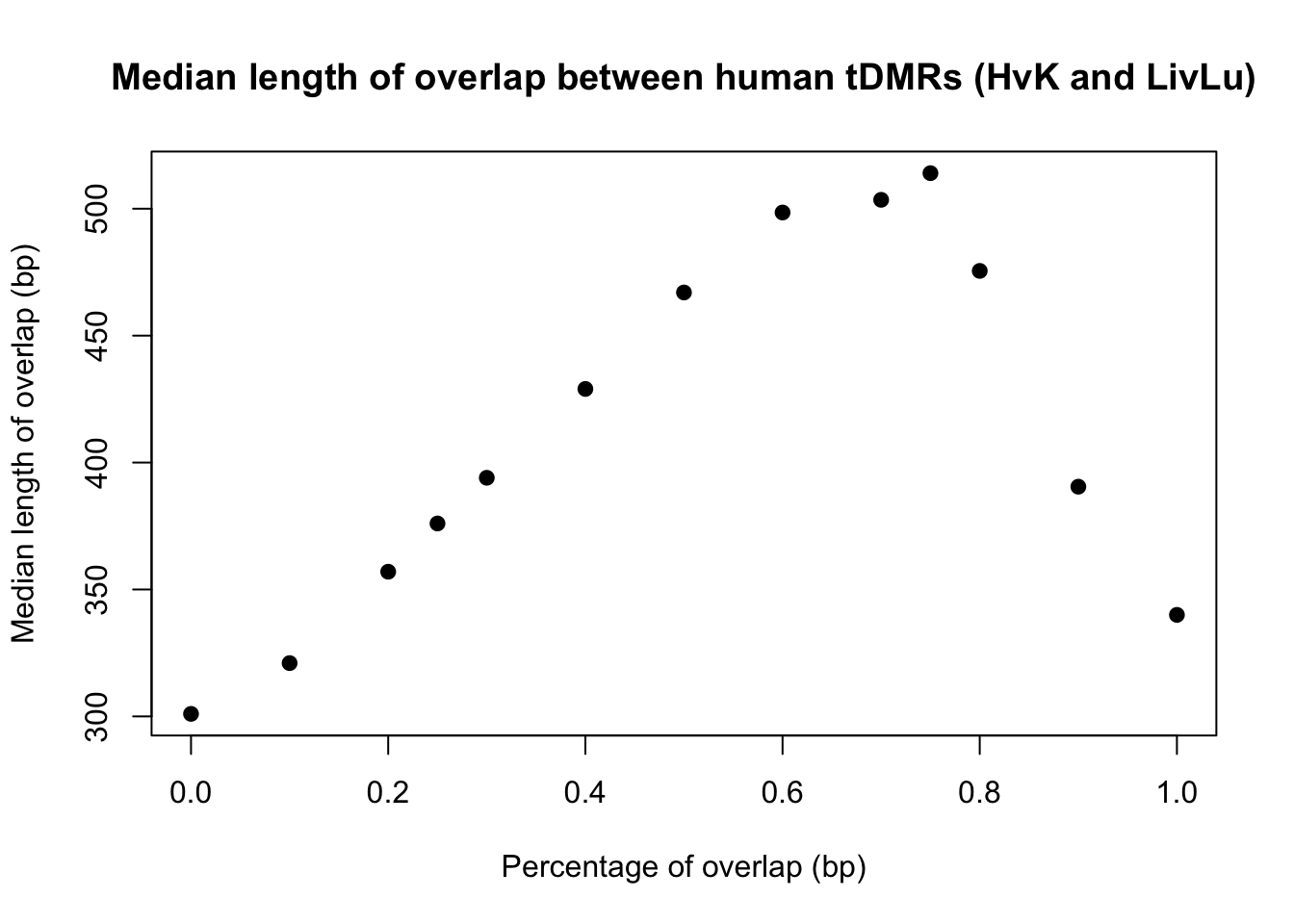
Are the overlap % results robust with respect to species?
Chimp heart versus liver and chimp heart versus lung
# columns to pull
pull_col <- c(1:3, 16)
# make bed style format: change name from human to rhesus
humans_heart_liver_DMRs_bed <- chimps_heart_liver_DMRs[,pull_col]
# Get DMRs on the autosomal chromosomes only
humans_heart_liver_DMRs_bed_noX <- humans_heart_liver_DMRs_bed[which(humans_heart_liver_DMRs_bed[,1] != "chrX"),]
humans_heart_liver_DMRs_bed_noX[,1] <- as.character(humans_heart_liver_DMRs_bed_noX[,1])
humans_heart_liver_DMRs_bed_noX[,2] <- as.integer(humans_heart_liver_DMRs_bed_noX[,2])
humans_heart_liver_DMRs_bed_noX[,3] <- as.integer(humans_heart_liver_DMRs_bed_noX[,3])
summary(humans_heart_liver_DMRs_bed_noX[,3] - humans_heart_liver_DMRs_bed_noX[,2]) Min. 1st Qu. Median Mean 3rd Qu. Max.
5.0 266.0 498.0 617.6 849.0 4950.0 sort_humans_heart_liver_DMRs_bed <- bedr.sort.region(humans_heart_liver_DMRs_bed_noX, check.chr = FALSE)SORTING
VALIDATE REGIONS
* Checking input type... PASS
Input is in bed format
* Check if index is a string... PASS
* Check index pattern... PASS
* Check for missing values... PASS
* Check for larger start position... PASS.
* Check if zero based... PASS
* Overlapping regions can cause unexpected results.# make bed style format
humans_heart_lung_DMRs_bed <- chimps_heart_lung_DMRs[,pull_col]
humans_heart_lung_DMRs_bed_noX <- humans_heart_lung_DMRs_bed[which(humans_heart_lung_DMRs_bed[,1] != "chrX"),]
humans_heart_lung_DMRs_bed_noX[,1] <- as.character(humans_heart_lung_DMRs_bed_noX[,1])
humans_heart_lung_DMRs_bed_noX[,2] <- as.integer(humans_heart_lung_DMRs_bed_noX[,2])
humans_heart_lung_DMRs_bed_noX[,3] <- as.integer(humans_heart_lung_DMRs_bed_noX[,3])
sort_humans_heart_lung_DMRs_bed <- bedr.sort.region(humans_heart_lung_DMRs_bed_noX, check.chr = FALSE)SORTING
VALIDATE REGIONS
* Checking input type... PASS
Input is in bed format
* Check if index is a string... PASS
* Check index pattern... PASS
* Check for missing values... PASS
* Check for larger start position... PASS.
* Check if zero based... PASS
* Overlapping regions can cause unexpected results.summary(humans_heart_lung_DMRs_bed_noX[,3] - humans_heart_lung_DMRs_bed_noX[,2]) Min. 1st Qu. Median Mean 3rd Qu. Max.
5.0 225.0 413.5 512.3 708.0 4186.0 heart_lung <- bedr(input = list(a = sort_humans_heart_liver_DMRs_bed, b = sort_humans_heart_lung_DMRs_bed), method = "intersect", params = "-wao -f 0.5 -F 0.5", check.chr = FALSE) * Processing input (1): a
CONVERT TO BED
* Checking input type... PASS
Input is in bed format
VALIDATE REGIONS
* Check if index is a string... PASS
* Check index pattern... PASS
* Check for missing values... PASS
* Check for larger start position... PASS.
* Check if zero based... PASS
* Checking sort order... PASS
* Checking for overlapping 'contiguous' regions... PASS
* Processing input (2): b
CONVERT TO BED
* Checking input type... PASS
Input is in bed format
VALIDATE REGIONS
* Check if index is a string... PASS
* Check index pattern... PASS
* Check for missing values... PASS
* Check for larger start position... PASS.
* Check if zero based... PASS
* Checking sort order... PASS
* Checking for overlapping 'contiguous' regions... PASS
bedtools intersect -a /var/folders/rf/qrcw6ncj05z1pc_pq9xzw3540000gn/T//Rtmph6wW6Y/a_c3e322cb955.bed -b /var/folders/rf/qrcw6ncj05z1pc_pq9xzw3540000gn/T//Rtmph6wW6Y/b_c3e341d5deed.bed -wao -f 0.5 -F 0.5summary(heart_lung[,3] - heart_lung[,2]) Min. 1st Qu. Median Mean 3rd Qu. Max.
5.0 266.0 498.0 617.6 849.0 4950.0 summary(as.integer(heart_lung[,9])) Min. 1st Qu. Median Mean 3rd Qu. Max.
0.00 0.00 0.00 56.97 0.00 4186.00 summary(as.integer(heart_lung[,9]) > 0) Mode FALSE TRUE
logical 26162 2607 lung_heart <- bedr(input = list(a = sort_humans_heart_lung_DMRs_bed, b = sort_humans_heart_liver_DMRs_bed), method = "intersect", params = "-wao -f 0.5 -F 0.5", check.chr = FALSE) * Processing input (1): a
CONVERT TO BED
* Checking input type... PASS
Input is in bed format
VALIDATE REGIONS
* Check if index is a string... PASS
* Check index pattern... PASS
* Check for missing values... PASS
* Check for larger start position... PASS.
* Check if zero based... PASS
* Checking sort order... PASS
* Checking for overlapping 'contiguous' regions... PASS
* Processing input (2): b
CONVERT TO BED
* Checking input type... PASS
Input is in bed format
VALIDATE REGIONS
* Check if index is a string... PASS
* Check index pattern... PASS
* Check for missing values... PASS
* Check for larger start position... PASS.
* Check if zero based... PASS
* Checking sort order... PASS
* Checking for overlapping 'contiguous' regions... PASS
bedtools intersect -a /var/folders/rf/qrcw6ncj05z1pc_pq9xzw3540000gn/T//Rtmph6wW6Y/a_c3e36d3b12c2.bed -b /var/folders/rf/qrcw6ncj05z1pc_pq9xzw3540000gn/T//Rtmph6wW6Y/b_c3e342e44a79.bed -wao -f 0.5 -F 0.5summary(as.integer(lung_heart[,9])) Min. 1st Qu. Median Mean 3rd Qu. Max.
0.0 0.0 0.0 192.2 278.8 4186.0 summary(as.integer(lung_heart[,9]) > 0) Mode FALSE TRUE
logical 5919 2607 # Make a Venn diagram based on the results
summary(as.integer(heart_lung[,9]) > 0) Mode FALSE TRUE
logical 26162 2607 summary(as.integer(lung_heart[,9]) > 0) Mode FALSE TRUE
logical 5919 2607 grid.newpage()
draw.pairwise.venn(area1 = (26162+2607), area2 = (5919+2607), cross.area = 2607, category = c("Heart v. Liver", "Heart v. Lung"))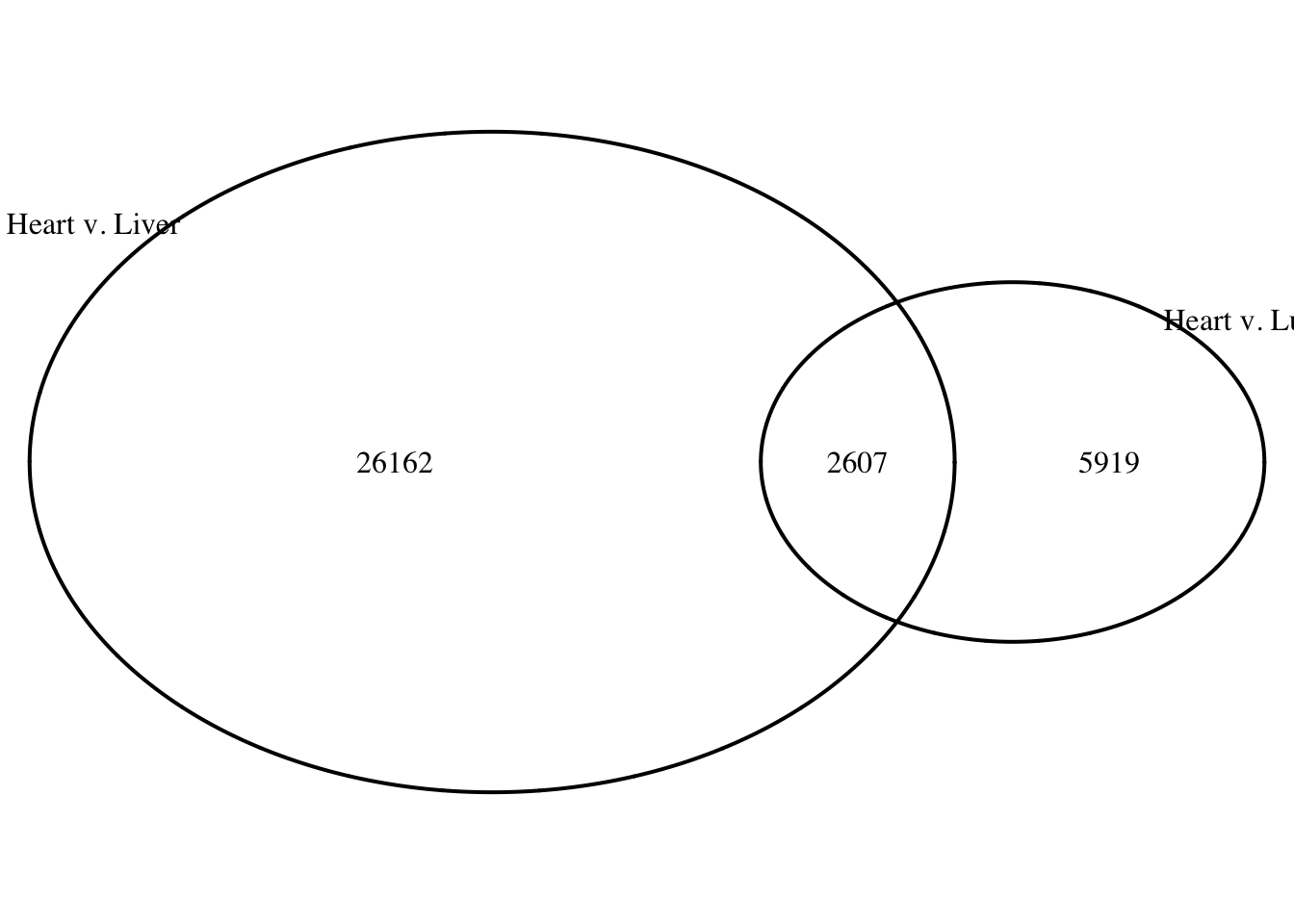
(polygon[GRID.polygon.20], polygon[GRID.polygon.21], polygon[GRID.polygon.22], polygon[GRID.polygon.23], text[GRID.text.24], text[GRID.text.25], text[GRID.text.26], text[GRID.text.27], text[GRID.text.28]) # 1 bp overlap
heart_liver_1 <- bedr(input = list(a = sort_humans_heart_liver_DMRs_bed, b = sort_humans_heart_lung_DMRs_bed), method = "intersect", params = "-wao -f 0.00000001 -F 0.00000001", check.chr = FALSE) * Processing input (1): a
CONVERT TO BED
* Checking input type... PASS
Input is in bed format
VALIDATE REGIONS
* Check if index is a string... PASS
* Check index pattern... PASS
* Check for missing values... PASS
* Check for larger start position... PASS.
* Check if zero based... PASS
* Checking sort order... PASS
* Checking for overlapping 'contiguous' regions... PASS
* Processing input (2): b
CONVERT TO BED
* Checking input type... PASS
Input is in bed format
VALIDATE REGIONS
* Check if index is a string... PASS
* Check index pattern... PASS
* Check for missing values... PASS
* Check for larger start position... PASS.
* Check if zero based... PASS
* Checking sort order... PASS
* Checking for overlapping 'contiguous' regions... PASS
bedtools intersect -a /var/folders/rf/qrcw6ncj05z1pc_pq9xzw3540000gn/T//Rtmph6wW6Y/a_c3e35d2cf2c5.bed -b /var/folders/rf/qrcw6ncj05z1pc_pq9xzw3540000gn/T//Rtmph6wW6Y/b_c3e35f708f26.bed -wao -f 0.00000001 -F 0.00000001# 10% overlap
heart_liver_10 <- bedr(input = list(a = sort_humans_heart_liver_DMRs_bed, b = sort_humans_heart_lung_DMRs_bed), method = "intersect", params = "-wao -f 0.10 -F 0.10", check.chr = FALSE) * Processing input (1): a
CONVERT TO BED
* Checking input type... PASS
Input is in bed format
VALIDATE REGIONS
* Check if index is a string... PASS
* Check index pattern... PASS
* Check for missing values... PASS
* Check for larger start position... PASS.
* Check if zero based... PASS
* Checking sort order... PASS
* Checking for overlapping 'contiguous' regions... PASS
* Processing input (2): b
CONVERT TO BED
* Checking input type... PASS
Input is in bed format
VALIDATE REGIONS
* Check if index is a string... PASS
* Check index pattern... PASS
* Check for missing values... PASS
* Check for larger start position... PASS.
* Check if zero based... PASS
* Checking sort order... PASS
* Checking for overlapping 'contiguous' regions... PASS
bedtools intersect -a /var/folders/rf/qrcw6ncj05z1pc_pq9xzw3540000gn/T//Rtmph6wW6Y/a_c3e362de3e60.bed -b /var/folders/rf/qrcw6ncj05z1pc_pq9xzw3540000gn/T//Rtmph6wW6Y/b_c3e319fed68.bed -wao -f 0.10 -F 0.10summary(as.integer(heart_liver_10[,9]) > 0) Mode FALSE TRUE
logical 24943 3837 # 20% overlap
heart_liver_20 <- bedr(input = list(a = sort_humans_heart_liver_DMRs_bed, b = sort_humans_heart_lung_DMRs_bed), method = "intersect", params = "-wao -f 0.20 -F 0.20", check.chr = FALSE) * Processing input (1): a
CONVERT TO BED
* Checking input type... PASS
Input is in bed format
VALIDATE REGIONS
* Check if index is a string... PASS
* Check index pattern... PASS
* Check for missing values... PASS
* Check for larger start position... PASS.
* Check if zero based... PASS
* Checking sort order... PASS
* Checking for overlapping 'contiguous' regions... PASS
* Processing input (2): b
CONVERT TO BED
* Checking input type... PASS
Input is in bed format
VALIDATE REGIONS
* Check if index is a string... PASS
* Check index pattern... PASS
* Check for missing values... PASS
* Check for larger start position... PASS.
* Check if zero based... PASS
* Checking sort order... PASS
* Checking for overlapping 'contiguous' regions... PASS
bedtools intersect -a /var/folders/rf/qrcw6ncj05z1pc_pq9xzw3540000gn/T//Rtmph6wW6Y/a_c3e3aa4640d.bed -b /var/folders/rf/qrcw6ncj05z1pc_pq9xzw3540000gn/T//Rtmph6wW6Y/b_c3e365e71b74.bed -wao -f 0.20 -F 0.20summary(as.integer(heart_liver_20[,9]) > 0) Mode FALSE TRUE
logical 25160 3614 # 25% overlap
heart_liver_25 <- bedr(input = list(a = sort_humans_heart_liver_DMRs_bed, b = sort_humans_heart_lung_DMRs_bed), method = "intersect", params = "-wao -f 0.25 -F 0.25", check.chr = FALSE) * Processing input (1): a
CONVERT TO BED
* Checking input type... PASS
Input is in bed format
VALIDATE REGIONS
* Check if index is a string... PASS
* Check index pattern... PASS
* Check for missing values... PASS
* Check for larger start position... PASS.
* Check if zero based... PASS
* Checking sort order... PASS
* Checking for overlapping 'contiguous' regions... PASS
* Processing input (2): b
CONVERT TO BED
* Checking input type... PASS
Input is in bed format
VALIDATE REGIONS
* Check if index is a string... PASS
* Check index pattern... PASS
* Check for missing values... PASS
* Check for larger start position... PASS.
* Check if zero based... PASS
* Checking sort order... PASS
* Checking for overlapping 'contiguous' regions... PASS
bedtools intersect -a /var/folders/rf/qrcw6ncj05z1pc_pq9xzw3540000gn/T//Rtmph6wW6Y/a_c3e31443c3f0.bed -b /var/folders/rf/qrcw6ncj05z1pc_pq9xzw3540000gn/T//Rtmph6wW6Y/b_c3e354f63da3.bed -wao -f 0.25 -F 0.25summary(as.integer(heart_liver_25[,9]) > 0) Mode FALSE TRUE
logical 25301 3472 # 30% overlap
heart_liver_30 <- bedr(input = list(a = sort_humans_heart_liver_DMRs_bed, b = sort_humans_heart_lung_DMRs_bed), method = "intersect", params = "-wao -f 0.30 -F 0.30", check.chr = FALSE) * Processing input (1): a
CONVERT TO BED
* Checking input type... PASS
Input is in bed format
VALIDATE REGIONS
* Check if index is a string... PASS
* Check index pattern... PASS
* Check for missing values... PASS
* Check for larger start position... PASS.
* Check if zero based... PASS
* Checking sort order... PASS
* Checking for overlapping 'contiguous' regions... PASS
* Processing input (2): b
CONVERT TO BED
* Checking input type... PASS
Input is in bed format
VALIDATE REGIONS
* Check if index is a string... PASS
* Check index pattern... PASS
* Check for missing values... PASS
* Check for larger start position... PASS.
* Check if zero based... PASS
* Checking sort order... PASS
* Checking for overlapping 'contiguous' regions... PASS
bedtools intersect -a /var/folders/rf/qrcw6ncj05z1pc_pq9xzw3540000gn/T//Rtmph6wW6Y/a_c3e365db5c2c.bed -b /var/folders/rf/qrcw6ncj05z1pc_pq9xzw3540000gn/T//Rtmph6wW6Y/b_c3e369495f96.bed -wao -f 0.30 -F 0.30summary(as.integer(heart_liver_30[,9]) > 0) Mode FALSE TRUE
logical 25439 3332 #40% overlap
heart_liver_40 <- bedr(input = list(a = sort_humans_heart_liver_DMRs_bed, b = sort_humans_heart_lung_DMRs_bed), method = "intersect", params = "-wao -f 0.4 -F 0.4", check.chr = FALSE) * Processing input (1): a
CONVERT TO BED
* Checking input type... PASS
Input is in bed format
VALIDATE REGIONS
* Check if index is a string... PASS
* Check index pattern... PASS
* Check for missing values... PASS
* Check for larger start position... PASS.
* Check if zero based... PASS
* Checking sort order... PASS
* Checking for overlapping 'contiguous' regions... PASS
* Processing input (2): b
CONVERT TO BED
* Checking input type... PASS
Input is in bed format
VALIDATE REGIONS
* Check if index is a string... PASS
* Check index pattern... PASS
* Check for missing values... PASS
* Check for larger start position... PASS.
* Check if zero based... PASS
* Checking sort order... PASS
* Checking for overlapping 'contiguous' regions... PASS
bedtools intersect -a /var/folders/rf/qrcw6ncj05z1pc_pq9xzw3540000gn/T//Rtmph6wW6Y/a_c3e36f32eaea.bed -b /var/folders/rf/qrcw6ncj05z1pc_pq9xzw3540000gn/T//Rtmph6wW6Y/b_c3e3794a197e.bed -wao -f 0.4 -F 0.4summary(as.integer(heart_liver_40[,9]) > 0) Mode FALSE TRUE
logical 25795 2974 # 50% overlap
heart_liver_50 <- bedr(input = list(a = sort_humans_heart_liver_DMRs_bed, b = sort_humans_heart_lung_DMRs_bed), method = "intersect", params = "-wao -f 0.50 -F 0.50", check.chr = FALSE) * Processing input (1): a
CONVERT TO BED
* Checking input type... PASS
Input is in bed format
VALIDATE REGIONS
* Check if index is a string... PASS
* Check index pattern... PASS
* Check for missing values... PASS
* Check for larger start position... PASS.
* Check if zero based... PASS
* Checking sort order... PASS
* Checking for overlapping 'contiguous' regions... PASS
* Processing input (2): b
CONVERT TO BED
* Checking input type... PASS
Input is in bed format
VALIDATE REGIONS
* Check if index is a string... PASS
* Check index pattern... PASS
* Check for missing values... PASS
* Check for larger start position... PASS.
* Check if zero based... PASS
* Checking sort order... PASS
* Checking for overlapping 'contiguous' regions... PASS
bedtools intersect -a /var/folders/rf/qrcw6ncj05z1pc_pq9xzw3540000gn/T//Rtmph6wW6Y/a_c3e36da7e2a6.bed -b /var/folders/rf/qrcw6ncj05z1pc_pq9xzw3540000gn/T//Rtmph6wW6Y/b_c3e3571beacc.bed -wao -f 0.50 -F 0.50summary(as.integer(heart_liver_50[,9]) > 0) Mode FALSE TRUE
logical 26162 2607 #60% overlap
heart_liver_60 <- bedr(input = list(a = sort_humans_heart_liver_DMRs_bed, b = sort_humans_heart_lung_DMRs_bed), method = "intersect", params = "-wao -f 0.6 -F 0.6", check.chr = FALSE) * Processing input (1): a
CONVERT TO BED
* Checking input type... PASS
Input is in bed format
VALIDATE REGIONS
* Check if index is a string... PASS
* Check index pattern... PASS
* Check for missing values... PASS
* Check for larger start position... PASS.
* Check if zero based... PASS
* Checking sort order... PASS
* Checking for overlapping 'contiguous' regions... PASS
* Processing input (2): b
CONVERT TO BED
* Checking input type... PASS
Input is in bed format
VALIDATE REGIONS
* Check if index is a string... PASS
* Check index pattern... PASS
* Check for missing values... PASS
* Check for larger start position... PASS.
* Check if zero based... PASS
* Checking sort order... PASS
* Checking for overlapping 'contiguous' regions... PASS
bedtools intersect -a /var/folders/rf/qrcw6ncj05z1pc_pq9xzw3540000gn/T//Rtmph6wW6Y/a_c3e332110690.bed -b /var/folders/rf/qrcw6ncj05z1pc_pq9xzw3540000gn/T//Rtmph6wW6Y/b_c3e3379becef.bed -wao -f 0.6 -F 0.6summary(as.integer(heart_liver_60[,9]) > 0) Mode FALSE TRUE
logical 26589 2180 # 70% overlap
heart_liver_70 <- bedr(input = list(a = sort_humans_heart_liver_DMRs_bed, b = sort_humans_heart_lung_DMRs_bed), method = "intersect", params = "-wao -f 0.70 -F 0.70", check.chr = FALSE) * Processing input (1): a
CONVERT TO BED
* Checking input type... PASS
Input is in bed format
VALIDATE REGIONS
* Check if index is a string... PASS
* Check index pattern... PASS
* Check for missing values... PASS
* Check for larger start position... PASS.
* Check if zero based... PASS
* Checking sort order... PASS
* Checking for overlapping 'contiguous' regions... PASS
* Processing input (2): b
CONVERT TO BED
* Checking input type... PASS
Input is in bed format
VALIDATE REGIONS
* Check if index is a string... PASS
* Check index pattern... PASS
* Check for missing values... PASS
* Check for larger start position... PASS.
* Check if zero based... PASS
* Checking sort order... PASS
* Checking for overlapping 'contiguous' regions... PASS
bedtools intersect -a /var/folders/rf/qrcw6ncj05z1pc_pq9xzw3540000gn/T//Rtmph6wW6Y/a_c3e3f3a284f.bed -b /var/folders/rf/qrcw6ncj05z1pc_pq9xzw3540000gn/T//Rtmph6wW6Y/b_c3e3775bb6e7.bed -wao -f 0.70 -F 0.70summary(as.integer(heart_liver_70[,9]) > 0) Mode FALSE TRUE
logical 27029 1740 #75% overlap
heart_liver_75 <- bedr(input = list(a = sort_humans_heart_liver_DMRs_bed, b = sort_humans_heart_lung_DMRs_bed), method = "intersect", params = "-wao -f 0.75 -F 0.75", check.chr = FALSE) * Processing input (1): a
CONVERT TO BED
* Checking input type... PASS
Input is in bed format
VALIDATE REGIONS
* Check if index is a string... PASS
* Check index pattern... PASS
* Check for missing values... PASS
* Check for larger start position... PASS.
* Check if zero based... PASS
* Checking sort order... PASS
* Checking for overlapping 'contiguous' regions... PASS
* Processing input (2): b
CONVERT TO BED
* Checking input type... PASS
Input is in bed format
VALIDATE REGIONS
* Check if index is a string... PASS
* Check index pattern... PASS
* Check for missing values... PASS
* Check for larger start position... PASS.
* Check if zero based... PASS
* Checking sort order... PASS
* Checking for overlapping 'contiguous' regions... PASS
bedtools intersect -a /var/folders/rf/qrcw6ncj05z1pc_pq9xzw3540000gn/T//Rtmph6wW6Y/a_c3e34f24d4a.bed -b /var/folders/rf/qrcw6ncj05z1pc_pq9xzw3540000gn/T//Rtmph6wW6Y/b_c3e3f40eca.bed -wao -f 0.75 -F 0.75summary(as.integer(heart_liver_75[,9]) > 0) Mode FALSE TRUE
logical 27267 1502 # 80% overlap
heart_liver_80 <- bedr(input = list(a = sort_humans_heart_liver_DMRs_bed, b = sort_humans_heart_lung_DMRs_bed), method = "intersect", params = "-wao -f 0.80 -F 0.80", check.chr = FALSE) * Processing input (1): a
CONVERT TO BED
* Checking input type... PASS
Input is in bed format
VALIDATE REGIONS
* Check if index is a string... PASS
* Check index pattern... PASS
* Check for missing values... PASS
* Check for larger start position... PASS.
* Check if zero based... PASS
* Checking sort order... PASS
* Checking for overlapping 'contiguous' regions... PASS
* Processing input (2): b
CONVERT TO BED
* Checking input type... PASS
Input is in bed format
VALIDATE REGIONS
* Check if index is a string... PASS
* Check index pattern... PASS
* Check for missing values... PASS
* Check for larger start position... PASS.
* Check if zero based... PASS
* Checking sort order... PASS
* Checking for overlapping 'contiguous' regions... PASS
bedtools intersect -a /var/folders/rf/qrcw6ncj05z1pc_pq9xzw3540000gn/T//Rtmph6wW6Y/a_c3e31beeaa88.bed -b /var/folders/rf/qrcw6ncj05z1pc_pq9xzw3540000gn/T//Rtmph6wW6Y/b_c3e3435021e.bed -wao -f 0.80 -F 0.80summary(as.integer(heart_liver_80[,9]) > 0) Mode FALSE TRUE
logical 27515 1254 #90% overlap
heart_liver_90 <- bedr(input = list(a = sort_humans_heart_liver_DMRs_bed, b = sort_humans_heart_lung_DMRs_bed), method = "intersect", params = "-wao -f 0.9 -F 0.9", check.chr = FALSE) * Processing input (1): a
CONVERT TO BED
* Checking input type... PASS
Input is in bed format
VALIDATE REGIONS
* Check if index is a string... PASS
* Check index pattern... PASS
* Check for missing values... PASS
* Check for larger start position... PASS.
* Check if zero based... PASS
* Checking sort order... PASS
* Checking for overlapping 'contiguous' regions... PASS
* Processing input (2): b
CONVERT TO BED
* Checking input type... PASS
Input is in bed format
VALIDATE REGIONS
* Check if index is a string... PASS
* Check index pattern... PASS
* Check for missing values... PASS
* Check for larger start position... PASS.
* Check if zero based... PASS
* Checking sort order... PASS
* Checking for overlapping 'contiguous' regions... PASS
bedtools intersect -a /var/folders/rf/qrcw6ncj05z1pc_pq9xzw3540000gn/T//Rtmph6wW6Y/a_c3e374fabe7b.bed -b /var/folders/rf/qrcw6ncj05z1pc_pq9xzw3540000gn/T//Rtmph6wW6Y/b_c3e342d70593.bed -wao -f 0.9 -F 0.9summary(as.integer(heart_liver_90[,9]) > 0) Mode FALSE TRUE
logical 27915 854 #100% overlap
heart_liver_100 <- bedr(input = list(a = sort_humans_heart_liver_DMRs_bed, b = sort_humans_heart_lung_DMRs_bed), method = "intersect", params = "-wao -f 1 -F 1", check.chr = FALSE) * Processing input (1): a
CONVERT TO BED
* Checking input type... PASS
Input is in bed format
VALIDATE REGIONS
* Check if index is a string... PASS
* Check index pattern... PASS
* Check for missing values... PASS
* Check for larger start position... PASS.
* Check if zero based... PASS
* Checking sort order... PASS
* Checking for overlapping 'contiguous' regions... PASS
* Processing input (2): b
CONVERT TO BED
* Checking input type... PASS
Input is in bed format
VALIDATE REGIONS
* Check if index is a string... PASS
* Check index pattern... PASS
* Check for missing values... PASS
* Check for larger start position... PASS.
* Check if zero based... PASS
* Checking sort order... PASS
* Checking for overlapping 'contiguous' regions... PASS
bedtools intersect -a /var/folders/rf/qrcw6ncj05z1pc_pq9xzw3540000gn/T//Rtmph6wW6Y/a_c3e36e98ab75.bed -b /var/folders/rf/qrcw6ncj05z1pc_pq9xzw3540000gn/T//Rtmph6wW6Y/b_c3e316d02663.bed -wao -f 1 -F 1summary(as.integer(heart_liver_100[,9]) > 0) Mode FALSE TRUE
logical 28148 621 # Find the number of tDMRs depending on the bp overlap
summary(as.integer(heart_liver_1[,9]) > 0) Mode FALSE TRUE
logical 24854 3940 summary(as.integer(heart_liver_10[,9]) > 0) Mode FALSE TRUE
logical 24943 3837 summary(as.integer(heart_liver_20[,9]) > 0) Mode FALSE TRUE
logical 25160 3614 summary(as.integer(heart_liver_25[,9]) > 0) Mode FALSE TRUE
logical 25301 3472 summary(as.integer(heart_liver_30[,9]) > 0) Mode FALSE TRUE
logical 25439 3332 summary(as.integer(heart_liver_40[,9]) > 0) Mode FALSE TRUE
logical 25795 2974 summary(as.integer(heart_liver_50[,9]) > 0) Mode FALSE TRUE
logical 26162 2607 summary(as.integer(heart_liver_60[,9]) > 0) Mode FALSE TRUE
logical 26589 2180 summary(as.integer(heart_liver_70[,9]) > 0) Mode FALSE TRUE
logical 27029 1740 summary(as.integer(heart_liver_75[,9]) > 0) Mode FALSE TRUE
logical 27267 1502 summary(as.integer(heart_liver_80[,9]) > 0) Mode FALSE TRUE
logical 27515 1254 summary(as.integer(heart_liver_90[,9]) > 0) Mode FALSE TRUE
logical 27915 854 summary(as.integer(heart_liver_100[,9]) > 0) Mode FALSE TRUE
logical 28148 621 # Plot the number of tDMRs
conserved_tdmr_heart_liver_heart_lung <- c(621, 3837, 3614, 3472, 3332, 2974, 2607, 2180, 1740, 1502,1254,854, 621)
perc_overlap <- c(0.00000001, 0.1, 0.2, 0.25, 0.3, 0.4, 0.5, 0.6, 0.7, 0.75, 0.80, 0.90, 1)
plot(perc_overlap, conserved_tdmr_heart_liver_heart_lung, xlab = "Percentage of overlap (bp)", ylab = "Number of overlapping chimp tDMRs", pch = 19, main = "Overlapping Chimp tDMRs (HvLi and HvLu)")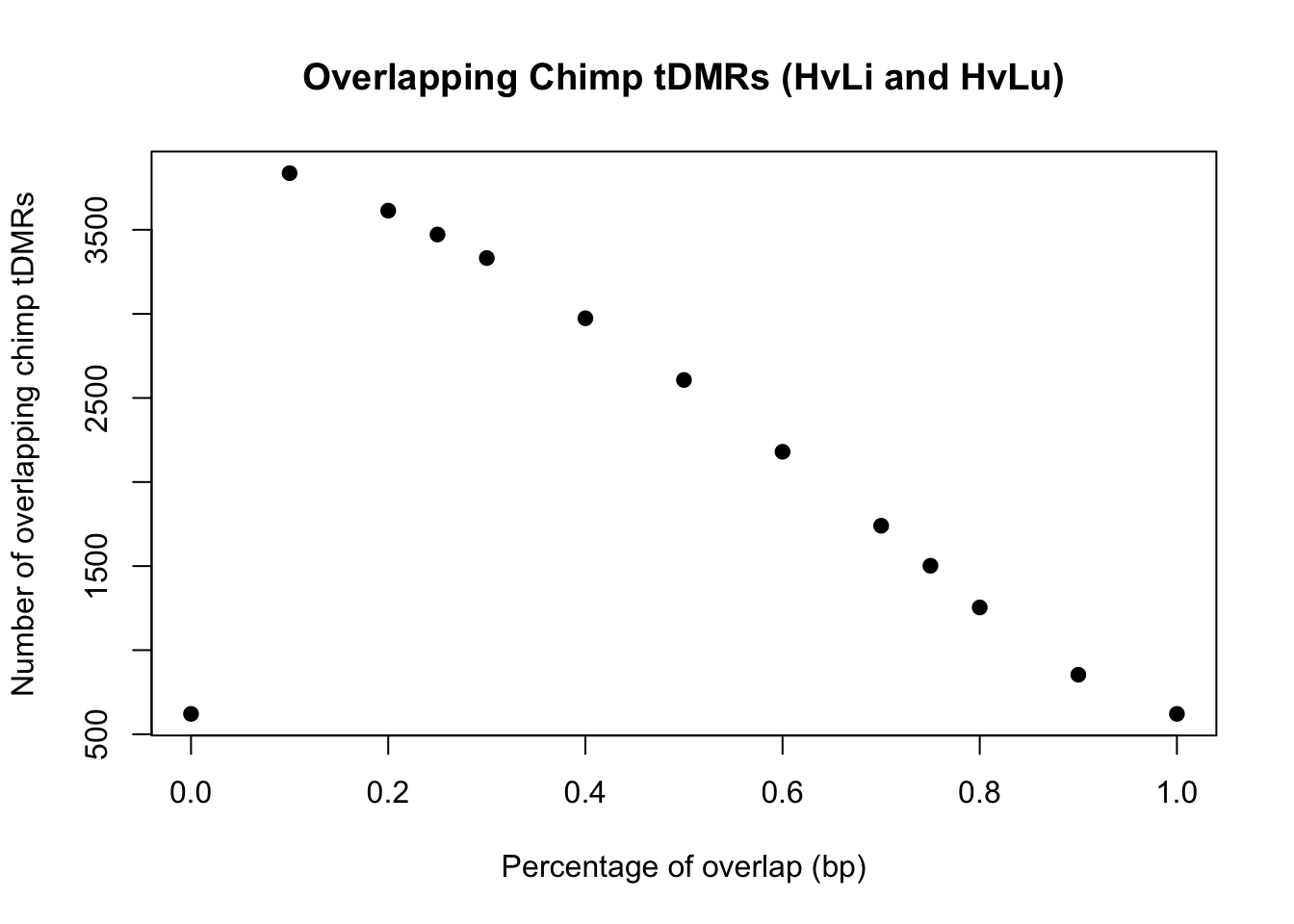
# Investigate the size of the tDMRs depending on the percentage overlap
check_1 <- heart_liver_1[which((as.integer(heart_liver_1[,9]) > 0) == TRUE),]
check_10 <- heart_liver_10[which((as.integer(heart_liver_10[,9]) > 0) == TRUE),]
check_20 <- heart_liver_20[which((as.integer(heart_liver_20[,9]) > 0) == TRUE),]
check_25 <- heart_liver_25[which((as.integer(heart_liver_25[,9]) > 0) == TRUE),]
check_30 <- heart_liver_30[which((as.integer(heart_liver_30[,9]) > 0) == TRUE),]
check_40 <- heart_liver_40[which((as.integer(heart_liver_40[,9]) > 0) == TRUE),]
check_50 <- heart_liver_50[which((as.integer(heart_liver_50[,9]) > 0) == TRUE),]
check_60 <- heart_liver_60[which((as.integer(heart_liver_60[,9]) > 0) == TRUE),]
check_70 <- heart_liver_70[which((as.integer(heart_liver_70[,9]) > 0) == TRUE),]
check_75 <- heart_liver_75[which((as.integer(heart_liver_75[,9]) > 0) == TRUE),]
check_80 <- heart_liver_80[which((as.integer(heart_liver_80[,9]) > 0) == TRUE),]
check_90 <- heart_liver_90[which((as.integer(heart_liver_90[,9]) > 0) == TRUE),]
check_100 <- heart_liver_100[which((as.integer(heart_liver_100[,9]) > 0) == TRUE),]
summary(as.integer(check_1[,9])) Min. 1st Qu. Median Mean 3rd Qu. Max.
5.0 228.0 423.5 509.1 703.0 4186.0 summary(as.integer(check_10[,9])) Min. 1st Qu. Median Mean 3rd Qu. Max.
9.0 242.0 433.0 520.8 709.0 4186.0 summary(as.integer(check_20[,9])) Min. 1st Qu. Median Mean 3rd Qu. Max.
15.0 270.0 459.5 544.2 733.0 4186.0 summary(as.integer(check_25[,9])) Min. 1st Qu. Median Mean 3rd Qu. Max.
15.0 280.0 477.5 557.2 745.0 4186.0 summary(as.integer(check_30[,9])) Min. 1st Qu. Median Mean 3rd Qu. Max.
15.0 291.0 491.0 570.1 761.2 4186.0 summary(as.integer(check_40[,9])) Min. 1st Qu. Median Mean 3rd Qu. Max.
15.0 316.0 533.5 599.8 793.0 4186.0 summary(as.integer(check_50[,9])) Min. 1st Qu. Median Mean 3rd Qu. Max.
15.0 343.0 560.0 628.7 834.0 4186.0 summary(as.integer(check_60[,9])) Min. 1st Qu. Median Mean 3rd Qu. Max.
15.0 363.8 597.0 660.2 873.5 4186.0 summary(as.integer(check_70[,9])) Min. 1st Qu. Median Mean 3rd Qu. Max.
15.0 361.0 608.0 678.2 911.0 4186.0 summary(as.integer(check_75[,9])) Min. 1st Qu. Median Mean 3rd Qu. Max.
15.0 351.2 612.0 679.6 915.8 4186.0 summary(as.integer(check_80[,9])) Min. 1st Qu. Median Mean 3rd Qu. Max.
15.0 327.8 589.0 677.2 921.0 4186.0 summary(as.integer(check_90[,9])) Min. 1st Qu. Median Mean 3rd Qu. Max.
15.0 277.2 537.5 626.8 863.8 4186.0 summary(as.integer(check_100[,9])) Min. 1st Qu. Median Mean 3rd Qu. Max.
15.0 230.0 428.0 528.9 740.0 2637.0 # Plot the median tDMR length
conserved_tdmr_heart_liver_heart_lung_bp <- c(301, 433, 459.5, 477.5, 491, 533.5, 560, 597, 608, 612,589,537.5, 428)
perc_overlap <- c(0.000000001, 0.1, 0.2, 0.25, 0.3, 0.4, 0.5, 0.6, 0.7, 0.75, 0.80, 0.90, 1)
plot(perc_overlap, conserved_tdmr_heart_liver_heart_lung_bp, xlab = "Percentage of overlap (bp)", ylab = "Median length of overlap (bp)", pch = 19, main = "Median length of overlap between chimp tDMRs (HvLi and HvLu)")
Rhesus heart versus liver and rhesus heart versus lung
# Heart versus liver
# columns to pull
pull_col <- c(1:3, 16)
# make bed style format: change name from human to rhesus
humans_heart_liver_DMRs_bed <- rhesus_heart_liver_DMRs[,pull_col]
# Remove chr17_gl000204_random
humans_heart_liver_DMRs_bed_noX <- humans_heart_liver_DMRs_bed[which(humans_heart_liver_DMRs_bed[,1] != "chr17_gl000204_random"),]
humans_heart_liver_DMRs_bed_noX[,1] <- as.character(humans_heart_liver_DMRs_bed_noX[,1])
humans_heart_liver_DMRs_bed_noX[,2] <- as.integer(humans_heart_liver_DMRs_bed_noX[,2])
humans_heart_liver_DMRs_bed_noX[,3] <- as.integer(humans_heart_liver_DMRs_bed_noX[,3])
# Remove chr11_gl000202_random
humans_heart_liver_DMRs_bed_noX <- humans_heart_liver_DMRs_bed_noX[which(humans_heart_liver_DMRs_bed_noX[,1] != "chr11_gl000202_random"),]
humans_heart_liver_DMRs_bed_noX[,1] <- as.character(humans_heart_liver_DMRs_bed_noX[,1])
humans_heart_liver_DMRs_bed_noX[,2] <- as.integer(humans_heart_liver_DMRs_bed_noX[,2])
humans_heart_liver_DMRs_bed_noX[,3] <- as.integer(humans_heart_liver_DMRs_bed_noX[,3])
# Get DMRs on the autosomal chromosomes only
humans_heart_liver_DMRs_bed_noX <- humans_heart_liver_DMRs_bed_noX[which(humans_heart_liver_DMRs_bed_noX[,1] != "chrX"),]
humans_heart_liver_DMRs_bed_noX[,1] <- as.character(humans_heart_liver_DMRs_bed_noX[,1])
humans_heart_liver_DMRs_bed_noX[,2] <- as.integer(humans_heart_liver_DMRs_bed_noX[,2])
humans_heart_liver_DMRs_bed_noX[,3] <- as.integer(humans_heart_liver_DMRs_bed_noX[,3])
summary(humans_heart_liver_DMRs_bed_noX[,3] - humans_heart_liver_DMRs_bed_noX[,2]) Min. 1st Qu. Median Mean 3rd Qu. Max.
4.0 277.0 524.0 658.6 906.0 6805.0 sort_humans_heart_liver_DMRs_bed <- bedr.sort.region(humans_heart_liver_DMRs_bed_noX, check.chr = FALSE)SORTING
VALIDATE REGIONS
* Checking input type... PASS
Input is in bed format
* Check if index is a string... PASS
* Check index pattern... PASS
* Check for missing values... PASS
* Check for larger start position... PASS.
* Check if zero based... PASS
* Overlapping regions can cause unexpected results.# Heart versus lung
humans_heart_lung_DMRs_bed <- rhesus_heart_lung_DMRs[,pull_col]
humans_heart_lung_DMRs_bed_noX <- humans_heart_lung_DMRs_bed[which(humans_heart_lung_DMRs_bed[,1] != "chr17_gl000204_random"),]
humans_heart_lung_DMRs_bed_noX[,1] <- as.character(humans_heart_lung_DMRs_bed_noX[,1])
humans_heart_lung_DMRs_bed_noX[,2] <- as.integer(humans_heart_lung_DMRs_bed_noX[,2])
humans_heart_lung_DMRs_bed_noX[,3] <- as.integer(humans_heart_lung_DMRs_bed_noX[,3])
humans_heart_lung_DMRs_bed_noX <- humans_heart_lung_DMRs_bed_noX[which(humans_heart_lung_DMRs_bed_noX[,1] != "chr11_gl000202_random"),]
humans_heart_lung_DMRs_bed_noX[,1] <- as.character(humans_heart_lung_DMRs_bed_noX[,1])
humans_heart_lung_DMRs_bed_noX[,2] <- as.integer(humans_heart_lung_DMRs_bed_noX[,2])
humans_heart_lung_DMRs_bed_noX[,3] <- as.integer(humans_heart_lung_DMRs_bed_noX[,3])
humans_heart_lung_DMRs_bed_noX <- humans_heart_lung_DMRs_bed_noX[which(humans_heart_lung_DMRs_bed_noX[,1] != "chrX"),]
humans_heart_lung_DMRs_bed_noX[,1] <- as.character(humans_heart_lung_DMRs_bed_noX[,1])
humans_heart_lung_DMRs_bed_noX[,2] <- as.integer(humans_heart_lung_DMRs_bed_noX[,2])
humans_heart_lung_DMRs_bed_noX[,3] <- as.integer(humans_heart_lung_DMRs_bed_noX[,3])
sort_humans_heart_lung_DMRs_bed <- bedr.sort.region(humans_heart_lung_DMRs_bed_noX, check.chr = FALSE)SORTING
VALIDATE REGIONS
* Checking input type... PASS
Input is in bed format
* Check if index is a string... PASS
* Check index pattern... PASS
* Check for missing values... PASS
* Check for larger start position... PASS.
* Check if zero based... PASS
* Overlapping regions can cause unexpected results.summary(humans_heart_lung_DMRs_bed_noX[,3] - humans_heart_lung_DMRs_bed_noX[,2]) Min. 1st Qu. Median Mean 3rd Qu. Max.
6.0 226.0 419.0 521.9 723.8 4002.0 heart_lung <- bedr(input = list(a = sort_humans_heart_liver_DMRs_bed, b = sort_humans_heart_lung_DMRs_bed), method = "intersect", params = "-wao -f 0.5 -F 0.5", check.chr = FALSE) * Processing input (1): a
CONVERT TO BED
* Checking input type... PASS
Input is in bed format
VALIDATE REGIONS
* Check if index is a string... PASS
* Check index pattern... PASS
* Check for missing values... PASS
* Check for larger start position... PASS.
* Check if zero based... PASS
* Checking sort order... PASS
* Checking for overlapping 'contiguous' regions... PASS
* Processing input (2): b
CONVERT TO BED
* Checking input type... PASS
Input is in bed format
VALIDATE REGIONS
* Check if index is a string... PASS
* Check index pattern... PASS
* Check for missing values... PASS
* Check for larger start position... PASS.
* Check if zero based... PASS
* Checking sort order... PASS
* Checking for overlapping 'contiguous' regions... PASS
bedtools intersect -a /var/folders/rf/qrcw6ncj05z1pc_pq9xzw3540000gn/T//Rtmph6wW6Y/a_c3e3529f0c7d.bed -b /var/folders/rf/qrcw6ncj05z1pc_pq9xzw3540000gn/T//Rtmph6wW6Y/b_c3e370fe6a48.bed -wao -f 0.5 -F 0.5summary(heart_lung[,3] - heart_lung[,2]) Min. 1st Qu. Median Mean 3rd Qu. Max.
4.0 277.0 524.0 658.6 906.0 6805.0 summary(as.integer(heart_lung[,9])) Min. 1st Qu. Median Mean 3rd Qu. Max.
0.00 0.00 0.00 35.26 0.00 3679.00 summary(as.integer(heart_lung[,9]) > 0) Mode FALSE TRUE
logical 39051 2229 lung_heart <- bedr(input = list(a = sort_humans_heart_lung_DMRs_bed, b = sort_humans_heart_liver_DMRs_bed), method = "intersect", params = "-wao -f 0.5 -F 0.5", check.chr = FALSE) * Processing input (1): a
CONVERT TO BED
* Checking input type... PASS
Input is in bed format
VALIDATE REGIONS
* Check if index is a string... PASS
* Check index pattern... PASS
* Check for missing values... PASS
* Check for larger start position... PASS.
* Check if zero based... PASS
* Checking sort order... PASS
* Checking for overlapping 'contiguous' regions... PASS
* Processing input (2): b
CONVERT TO BED
* Checking input type... PASS
Input is in bed format
VALIDATE REGIONS
* Check if index is a string... PASS
* Check index pattern... PASS
* Check for missing values... PASS
* Check for larger start position... PASS.
* Check if zero based... PASS
* Checking sort order... PASS
* Checking for overlapping 'contiguous' regions... PASS
bedtools intersect -a /var/folders/rf/qrcw6ncj05z1pc_pq9xzw3540000gn/T//Rtmph6wW6Y/a_c3e35e96b10.bed -b /var/folders/rf/qrcw6ncj05z1pc_pq9xzw3540000gn/T//Rtmph6wW6Y/b_c3e363b20c8e.bed -wao -f 0.5 -F 0.5summary(as.integer(lung_heart[,9])) Min. 1st Qu. Median Mean 3rd Qu. Max.
0.0 0.0 0.0 207.1 302.8 3679.0 summary(as.integer(lung_heart[,9]) > 0) Mode FALSE TRUE
logical 4797 2229 # Make a Venn diagram based on the results
summary(as.integer(heart_lung[,9]) > 0) Mode FALSE TRUE
logical 39051 2229 summary(as.integer(lung_heart[,9]) > 0) Mode FALSE TRUE
logical 4797 2229 grid.newpage()
draw.pairwise.venn(area1 = (39057+2229), area2 = (4798+2229), cross.area = 2229, category = c("Heart v. Liver", "Heart v. Lung"))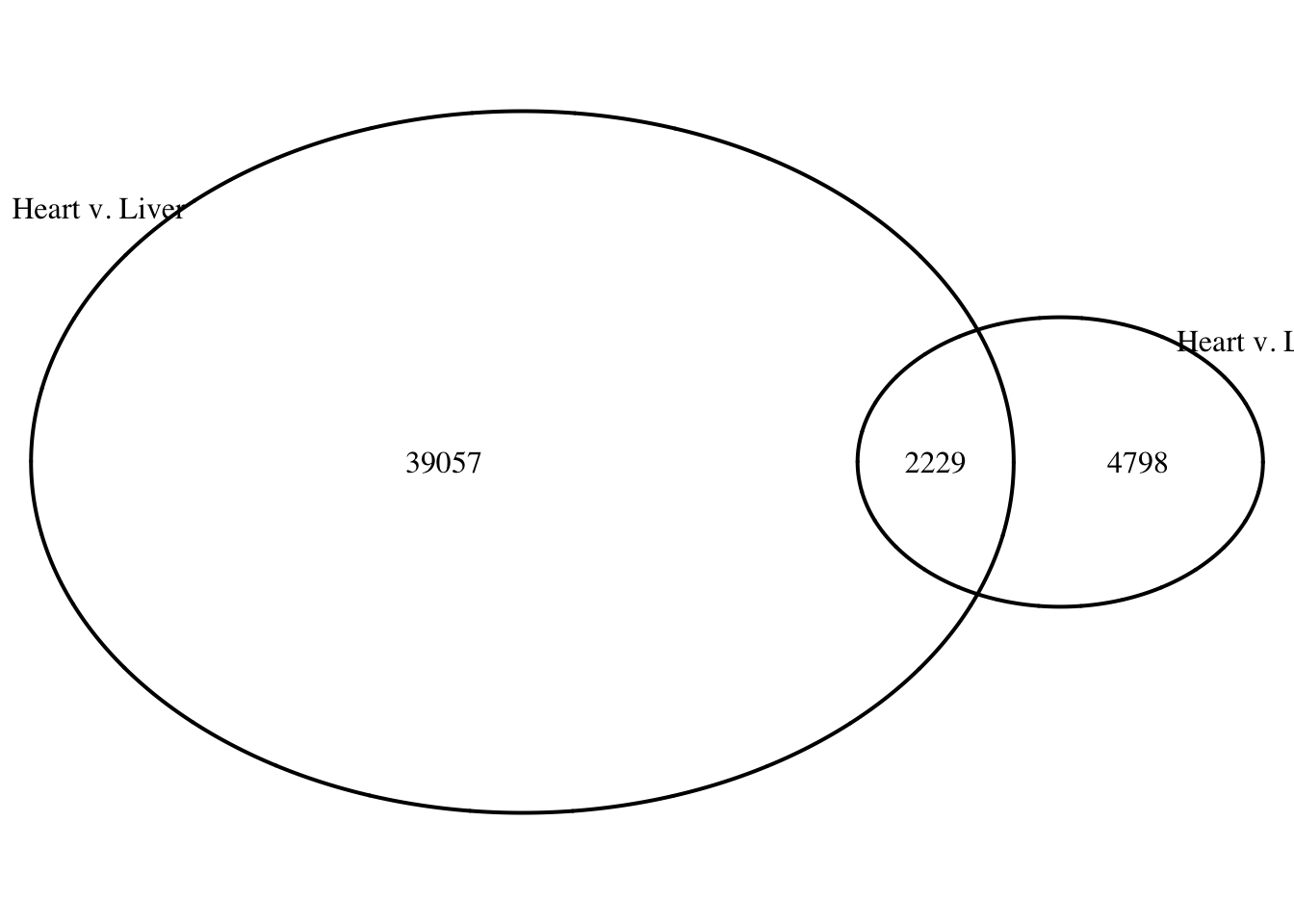
(polygon[GRID.polygon.29], polygon[GRID.polygon.30], polygon[GRID.polygon.31], polygon[GRID.polygon.32], text[GRID.text.33], text[GRID.text.34], text[GRID.text.35], text[GRID.text.36], text[GRID.text.37]) # 1 bp overlap
heart_liver_1 <- bedr(input = list(a = sort_humans_heart_liver_DMRs_bed, b = sort_humans_heart_lung_DMRs_bed), method = "intersect", params = "-wao -f 0.00000001 -F 0.00000001") * Processing input (1): a
CONVERT TO BED
* Checking input type... PASS
Input is in bed format
VALIDATE REGIONS
* Check if index is a string... PASS
* Check index pattern... PASS
* Check for missing values... PASS
* Check for larger start position... PASS.
* Check if zero based... PASS
* Checking sort order... PASS
* Checking for overlapping 'contiguous' regions... PASS
* Processing input (2): b
CONVERT TO BED
* Checking input type... PASS
Input is in bed format
VALIDATE REGIONS
* Check if index is a string... PASS
* Check index pattern... PASS
* Check for missing values... PASS
* Check for larger start position... PASS.
* Check if zero based... PASS
* Checking sort order... PASS
* Checking for overlapping 'contiguous' regions... PASS
bedtools intersect -a /var/folders/rf/qrcw6ncj05z1pc_pq9xzw3540000gn/T//Rtmph6wW6Y/a_c3e36dec683b.bed -b /var/folders/rf/qrcw6ncj05z1pc_pq9xzw3540000gn/T//Rtmph6wW6Y/b_c3e378a8089c.bed -wao -f 0.00000001 -F 0.00000001summary(as.integer(heart_liver_1[,9]) > 0) Mode FALSE TRUE
logical 37645 3654 # 10% overlap
heart_liver_10 <- bedr(input = list(a = sort_humans_heart_liver_DMRs_bed, b = sort_humans_heart_lung_DMRs_bed), method = "intersect", params = "-wao -f 0.10 -F 0.10", check.chr = FALSE) * Processing input (1): a
CONVERT TO BED
* Checking input type... PASS
Input is in bed format
VALIDATE REGIONS
* Check if index is a string... PASS
* Check index pattern... PASS
* Check for missing values... PASS
* Check for larger start position... PASS.
* Check if zero based... PASS
* Checking sort order... PASS
* Checking for overlapping 'contiguous' regions... PASS
* Processing input (2): b
CONVERT TO BED
* Checking input type... PASS
Input is in bed format
VALIDATE REGIONS
* Check if index is a string... PASS
* Check index pattern... PASS
* Check for missing values... PASS
* Check for larger start position... PASS.
* Check if zero based... PASS
* Checking sort order... PASS
* Checking for overlapping 'contiguous' regions... PASS
bedtools intersect -a /var/folders/rf/qrcw6ncj05z1pc_pq9xzw3540000gn/T//Rtmph6wW6Y/a_c3e32ee745dc.bed -b /var/folders/rf/qrcw6ncj05z1pc_pq9xzw3540000gn/T//Rtmph6wW6Y/b_c3e338aec9dd.bed -wao -f 0.10 -F 0.10summary(as.integer(heart_liver_10[,9]) > 0) Mode FALSE TRUE
logical 37776 3520 # 20% overlap
heart_liver_20 <- bedr(input = list(a = sort_humans_heart_liver_DMRs_bed, b = sort_humans_heart_lung_DMRs_bed), method = "intersect", params = "-wao -f 0.20 -F 0.20", check.chr = FALSE) * Processing input (1): a
CONVERT TO BED
* Checking input type... PASS
Input is in bed format
VALIDATE REGIONS
* Check if index is a string... PASS
* Check index pattern... PASS
* Check for missing values... PASS
* Check for larger start position... PASS.
* Check if zero based... PASS
* Checking sort order... PASS
* Checking for overlapping 'contiguous' regions... PASS
* Processing input (2): b
CONVERT TO BED
* Checking input type... PASS
Input is in bed format
VALIDATE REGIONS
* Check if index is a string... PASS
* Check index pattern... PASS
* Check for missing values... PASS
* Check for larger start position... PASS.
* Check if zero based... PASS
* Checking sort order... PASS
* Checking for overlapping 'contiguous' regions... PASS
bedtools intersect -a /var/folders/rf/qrcw6ncj05z1pc_pq9xzw3540000gn/T//Rtmph6wW6Y/a_c3e35399062a.bed -b /var/folders/rf/qrcw6ncj05z1pc_pq9xzw3540000gn/T//Rtmph6wW6Y/b_c3e3140b2be4.bed -wao -f 0.20 -F 0.20summary(as.integer(heart_liver_20[,9]) > 0) Mode FALSE TRUE
logical 38046 3245 # 25% overlap
heart_liver_25 <- bedr(input = list(a = sort_humans_heart_liver_DMRs_bed, b = sort_humans_heart_lung_DMRs_bed), method = "intersect", params = "-wao -f 0.25 -F 0.25", check.chr = FALSE) * Processing input (1): a
CONVERT TO BED
* Checking input type... PASS
Input is in bed format
VALIDATE REGIONS
* Check if index is a string... PASS
* Check index pattern... PASS
* Check for missing values... PASS
* Check for larger start position... PASS.
* Check if zero based... PASS
* Checking sort order... PASS
* Checking for overlapping 'contiguous' regions... PASS
* Processing input (2): b
CONVERT TO BED
* Checking input type... PASS
Input is in bed format
VALIDATE REGIONS
* Check if index is a string... PASS
* Check index pattern... PASS
* Check for missing values... PASS
* Check for larger start position... PASS.
* Check if zero based... PASS
* Checking sort order... PASS
* Checking for overlapping 'contiguous' regions... PASS
bedtools intersect -a /var/folders/rf/qrcw6ncj05z1pc_pq9xzw3540000gn/T//Rtmph6wW6Y/a_c3e36e168838.bed -b /var/folders/rf/qrcw6ncj05z1pc_pq9xzw3540000gn/T//Rtmph6wW6Y/b_c3e327d5f682.bed -wao -f 0.25 -F 0.25summary(as.integer(heart_liver_25[,9]) > 0) Mode FALSE TRUE
logical 38205 3084 # 30% overlap
heart_liver_30 <- bedr(input = list(a = sort_humans_heart_liver_DMRs_bed, b = sort_humans_heart_lung_DMRs_bed), method = "intersect", params = "-wao -f 0.30 -F 0.30", check.chr = FALSE) * Processing input (1): a
CONVERT TO BED
* Checking input type... PASS
Input is in bed format
VALIDATE REGIONS
* Check if index is a string... PASS
* Check index pattern... PASS
* Check for missing values... PASS
* Check for larger start position... PASS.
* Check if zero based... PASS
* Checking sort order... PASS
* Checking for overlapping 'contiguous' regions... PASS
* Processing input (2): b
CONVERT TO BED
* Checking input type... PASS
Input is in bed format
VALIDATE REGIONS
* Check if index is a string... PASS
* Check index pattern... PASS
* Check for missing values... PASS
* Check for larger start position... PASS.
* Check if zero based... PASS
* Checking sort order... PASS
* Checking for overlapping 'contiguous' regions... PASS
bedtools intersect -a /var/folders/rf/qrcw6ncj05z1pc_pq9xzw3540000gn/T//Rtmph6wW6Y/a_c3e31b04eec5.bed -b /var/folders/rf/qrcw6ncj05z1pc_pq9xzw3540000gn/T//Rtmph6wW6Y/b_c3e35ad8399a.bed -wao -f 0.30 -F 0.30summary(as.integer(heart_liver_30[,9]) > 0) Mode FALSE TRUE
logical 38353 2931 #40% overlap
heart_liver_40 <- bedr(input = list(a = sort_humans_heart_liver_DMRs_bed, b = sort_humans_heart_lung_DMRs_bed), method = "intersect", params = "-wao -f 0.4 -F 0.4", check.chr = FALSE) * Processing input (1): a
CONVERT TO BED
* Checking input type... PASS
Input is in bed format
VALIDATE REGIONS
* Check if index is a string... PASS
* Check index pattern... PASS
* Check for missing values... PASS
* Check for larger start position... PASS.
* Check if zero based... PASS
* Checking sort order... PASS
* Checking for overlapping 'contiguous' regions... PASS
* Processing input (2): b
CONVERT TO BED
* Checking input type... PASS
Input is in bed format
VALIDATE REGIONS
* Check if index is a string... PASS
* Check index pattern... PASS
* Check for missing values... PASS
* Check for larger start position... PASS.
* Check if zero based... PASS
* Checking sort order... PASS
* Checking for overlapping 'contiguous' regions... PASS
bedtools intersect -a /var/folders/rf/qrcw6ncj05z1pc_pq9xzw3540000gn/T//Rtmph6wW6Y/a_c3e379b6d28c.bed -b /var/folders/rf/qrcw6ncj05z1pc_pq9xzw3540000gn/T//Rtmph6wW6Y/b_c3e35f7a4e52.bed -wao -f 0.4 -F 0.4summary(as.integer(heart_liver_40[,9]) > 0) Mode FALSE TRUE
logical 38703 2577 # 50% overlap
heart_liver_50 <- bedr(input = list(a = sort_humans_heart_liver_DMRs_bed, b = sort_humans_heart_lung_DMRs_bed), method = "intersect", params = "-wao -f 0.50 -F 0.50", check.chr = FALSE) * Processing input (1): a
CONVERT TO BED
* Checking input type... PASS
Input is in bed format
VALIDATE REGIONS
* Check if index is a string... PASS
* Check index pattern... PASS
* Check for missing values... PASS
* Check for larger start position... PASS.
* Check if zero based... PASS
* Checking sort order... PASS
* Checking for overlapping 'contiguous' regions... PASS
* Processing input (2): b
CONVERT TO BED
* Checking input type... PASS
Input is in bed format
VALIDATE REGIONS
* Check if index is a string... PASS
* Check index pattern... PASS
* Check for missing values... PASS
* Check for larger start position... PASS.
* Check if zero based... PASS
* Checking sort order... PASS
* Checking for overlapping 'contiguous' regions... PASS
bedtools intersect -a /var/folders/rf/qrcw6ncj05z1pc_pq9xzw3540000gn/T//Rtmph6wW6Y/a_c3e3200e380a.bed -b /var/folders/rf/qrcw6ncj05z1pc_pq9xzw3540000gn/T//Rtmph6wW6Y/b_c3e32112e21f.bed -wao -f 0.50 -F 0.50summary(as.integer(heart_liver_50[,9]) > 0) Mode FALSE TRUE
logical 39051 2229 #60% overlap
heart_liver_60 <- bedr(input = list(a = sort_humans_heart_liver_DMRs_bed, b = sort_humans_heart_lung_DMRs_bed), method = "intersect", params = "-wao -f 0.6 -F 0.6", check.chr = FALSE) * Processing input (1): a
CONVERT TO BED
* Checking input type... PASS
Input is in bed format
VALIDATE REGIONS
* Check if index is a string... PASS
* Check index pattern... PASS
* Check for missing values... PASS
* Check for larger start position... PASS.
* Check if zero based... PASS
* Checking sort order... PASS
* Checking for overlapping 'contiguous' regions... PASS
* Processing input (2): b
CONVERT TO BED
* Checking input type... PASS
Input is in bed format
VALIDATE REGIONS
* Check if index is a string... PASS
* Check index pattern... PASS
* Check for missing values... PASS
* Check for larger start position... PASS.
* Check if zero based... PASS
* Checking sort order... PASS
* Checking for overlapping 'contiguous' regions... PASS
bedtools intersect -a /var/folders/rf/qrcw6ncj05z1pc_pq9xzw3540000gn/T//Rtmph6wW6Y/a_c3e316151f87.bed -b /var/folders/rf/qrcw6ncj05z1pc_pq9xzw3540000gn/T//Rtmph6wW6Y/b_c3e32346f519.bed -wao -f 0.6 -F 0.6summary(as.integer(heart_liver_60[,9]) > 0) Mode FALSE TRUE
logical 39421 1859 # 70% overlap
heart_liver_70 <- bedr(input = list(a = sort_humans_heart_liver_DMRs_bed, b = sort_humans_heart_lung_DMRs_bed), method = "intersect", params = "-wao -f 0.70 -F 0.70", check.chr = FALSE) * Processing input (1): a
CONVERT TO BED
* Checking input type... PASS
Input is in bed format
VALIDATE REGIONS
* Check if index is a string... PASS
* Check index pattern... PASS
* Check for missing values... PASS
* Check for larger start position... PASS.
* Check if zero based... PASS
* Checking sort order... PASS
* Checking for overlapping 'contiguous' regions... PASS
* Processing input (2): b
CONVERT TO BED
* Checking input type... PASS
Input is in bed format
VALIDATE REGIONS
* Check if index is a string... PASS
* Check index pattern... PASS
* Check for missing values... PASS
* Check for larger start position... PASS.
* Check if zero based... PASS
* Checking sort order... PASS
* Checking for overlapping 'contiguous' regions... PASS
bedtools intersect -a /var/folders/rf/qrcw6ncj05z1pc_pq9xzw3540000gn/T//Rtmph6wW6Y/a_c3e343596519.bed -b /var/folders/rf/qrcw6ncj05z1pc_pq9xzw3540000gn/T//Rtmph6wW6Y/b_c3e37e762ec4.bed -wao -f 0.70 -F 0.70summary(as.integer(heart_liver_70[,9]) > 0) Mode FALSE TRUE
logical 39802 1478 #75% overlap
heart_liver_75 <- bedr(input = list(a = sort_humans_heart_liver_DMRs_bed, b = sort_humans_heart_lung_DMRs_bed), method = "intersect", params = "-wao -f 0.75 -F 0.75", check.chr = FALSE) * Processing input (1): a
CONVERT TO BED
* Checking input type... PASS
Input is in bed format
VALIDATE REGIONS
* Check if index is a string... PASS
* Check index pattern... PASS
* Check for missing values... PASS
* Check for larger start position... PASS.
* Check if zero based... PASS
* Checking sort order... PASS
* Checking for overlapping 'contiguous' regions... PASS
* Processing input (2): b
CONVERT TO BED
* Checking input type... PASS
Input is in bed format
VALIDATE REGIONS
* Check if index is a string... PASS
* Check index pattern... PASS
* Check for missing values... PASS
* Check for larger start position... PASS.
* Check if zero based... PASS
* Checking sort order... PASS
* Checking for overlapping 'contiguous' regions... PASS
bedtools intersect -a /var/folders/rf/qrcw6ncj05z1pc_pq9xzw3540000gn/T//Rtmph6wW6Y/a_c3e367436ef3.bed -b /var/folders/rf/qrcw6ncj05z1pc_pq9xzw3540000gn/T//Rtmph6wW6Y/b_c3e31470110c.bed -wao -f 0.75 -F 0.75summary(as.integer(heart_liver_75[,9]) > 0) Mode FALSE TRUE
logical 39992 1288 # 80% overlap
heart_liver_80 <- bedr(input = list(a = sort_humans_heart_liver_DMRs_bed, b = sort_humans_heart_lung_DMRs_bed), method = "intersect", params = "-wao -f 0.80 -F 0.80", check.chr = FALSE) * Processing input (1): a
CONVERT TO BED
* Checking input type... PASS
Input is in bed format
VALIDATE REGIONS
* Check if index is a string... PASS
* Check index pattern... PASS
* Check for missing values... PASS
* Check for larger start position... PASS.
* Check if zero based... PASS
* Checking sort order... PASS
* Checking for overlapping 'contiguous' regions... PASS
* Processing input (2): b
CONVERT TO BED
* Checking input type... PASS
Input is in bed format
VALIDATE REGIONS
* Check if index is a string... PASS
* Check index pattern... PASS
* Check for missing values... PASS
* Check for larger start position... PASS.
* Check if zero based... PASS
* Checking sort order... PASS
* Checking for overlapping 'contiguous' regions... PASS
bedtools intersect -a /var/folders/rf/qrcw6ncj05z1pc_pq9xzw3540000gn/T//Rtmph6wW6Y/a_c3e35d77e3c6.bed -b /var/folders/rf/qrcw6ncj05z1pc_pq9xzw3540000gn/T//Rtmph6wW6Y/b_c3e320d3254a.bed -wao -f 0.80 -F 0.80summary(as.integer(heart_liver_80[,9]) > 0) Mode FALSE TRUE
logical 40179 1101 #90% overlap
heart_liver_90 <- bedr(input = list(a = sort_humans_heart_liver_DMRs_bed, b = sort_humans_heart_lung_DMRs_bed), method = "intersect", params = "-wao -f 0.9 -F 0.9", check.chr = FALSE) * Processing input (1): a
CONVERT TO BED
* Checking input type... PASS
Input is in bed format
VALIDATE REGIONS
* Check if index is a string... PASS
* Check index pattern... PASS
* Check for missing values... PASS
* Check for larger start position... PASS.
* Check if zero based... PASS
* Checking sort order... PASS
* Checking for overlapping 'contiguous' regions... PASS
* Processing input (2): b
CONVERT TO BED
* Checking input type... PASS
Input is in bed format
VALIDATE REGIONS
* Check if index is a string... PASS
* Check index pattern... PASS
* Check for missing values... PASS
* Check for larger start position... PASS.
* Check if zero based... PASS
* Checking sort order... PASS
* Checking for overlapping 'contiguous' regions... PASS
bedtools intersect -a /var/folders/rf/qrcw6ncj05z1pc_pq9xzw3540000gn/T//Rtmph6wW6Y/a_c3e330bbc9e4.bed -b /var/folders/rf/qrcw6ncj05z1pc_pq9xzw3540000gn/T//Rtmph6wW6Y/b_c3e371a217f8.bed -wao -f 0.9 -F 0.9summary(as.integer(heart_liver_90[,9]) > 0) Mode FALSE TRUE
logical 40542 738 #100% overlap
heart_liver_100 <- bedr(input = list(a = sort_humans_heart_liver_DMRs_bed, b = sort_humans_heart_lung_DMRs_bed), method = "intersect", params = "-wao -f 1 -F 1", check.chr = FALSE) * Processing input (1): a
CONVERT TO BED
* Checking input type... PASS
Input is in bed format
VALIDATE REGIONS
* Check if index is a string... PASS
* Check index pattern... PASS
* Check for missing values... PASS
* Check for larger start position... PASS.
* Check if zero based... PASS
* Checking sort order... PASS
* Checking for overlapping 'contiguous' regions... PASS
* Processing input (2): b
CONVERT TO BED
* Checking input type... PASS
Input is in bed format
VALIDATE REGIONS
* Check if index is a string... PASS
* Check index pattern... PASS
* Check for missing values... PASS
* Check for larger start position... PASS.
* Check if zero based... PASS
* Checking sort order... PASS
* Checking for overlapping 'contiguous' regions... PASS
bedtools intersect -a /var/folders/rf/qrcw6ncj05z1pc_pq9xzw3540000gn/T//Rtmph6wW6Y/a_c3e332a59263.bed -b /var/folders/rf/qrcw6ncj05z1pc_pq9xzw3540000gn/T//Rtmph6wW6Y/b_c3e317bd74d1.bed -wao -f 1 -F 1summary(as.integer(heart_liver_100[,9]) > 0) Mode FALSE TRUE
logical 40753 527 # Find the number of tDMRs depending on the bp overlap
summary(as.integer(heart_liver_1[,9]) > 0) Mode FALSE TRUE
logical 37645 3654 summary(as.integer(heart_liver_10[,9]) > 0) Mode FALSE TRUE
logical 37776 3520 summary(as.integer(heart_liver_20[,9]) > 0) Mode FALSE TRUE
logical 38046 3245 summary(as.integer(heart_liver_25[,9]) > 0) Mode FALSE TRUE
logical 38205 3084 summary(as.integer(heart_liver_30[,9]) > 0) Mode FALSE TRUE
logical 38353 2931 summary(as.integer(heart_liver_40[,9]) > 0) Mode FALSE TRUE
logical 38703 2577 summary(as.integer(heart_liver_50[,9]) > 0) Mode FALSE TRUE
logical 39051 2229 summary(as.integer(heart_liver_60[,9]) > 0) Mode FALSE TRUE
logical 39421 1859 summary(as.integer(heart_liver_70[,9]) > 0) Mode FALSE TRUE
logical 39802 1478 summary(as.integer(heart_liver_75[,9]) > 0) Mode FALSE TRUE
logical 39992 1288 summary(as.integer(heart_liver_80[,9]) > 0) Mode FALSE TRUE
logical 40179 1101 summary(as.integer(heart_liver_90[,9]) > 0) Mode FALSE TRUE
logical 40542 738 summary(as.integer(heart_liver_100[,9]) > 0) Mode FALSE TRUE
logical 40753 527 # Plot the number of tDMRs
conserved_tdmr_heart_liver_heart_lung <- c(3654, 3520, 3245, 3472, 3332, 2974, 2607, 2180, 1740, 1502,1254,854, 621)
perc_overlap <- c(0.000000001, 0.1, 0.2, 0.25, 0.3, 0.4, 0.5, 0.6, 0.7, 0.75, 0.80, 0.90, 1)
plot(perc_overlap, conserved_tdmr_heart_liver_heart_lung, xlab = "Percentage of overlap (bp)", ylab = "Number of overlapping rhesus tDMRs (HvLi and HvLu)", pch = 19)
# Investigate the size of the tDMRs depending on the percentage overlap
check_1 <- heart_liver_1[which((as.integer(heart_liver_1[,9]) > 0) == TRUE),]
check_10 <- heart_liver_10[which((as.integer(heart_liver_10[,9]) > 0) == TRUE),]
check_20 <- heart_liver_20[which((as.integer(heart_liver_20[,9]) > 0) == TRUE),]
check_25 <- heart_liver_25[which((as.integer(heart_liver_25[,9]) > 0) == TRUE),]
check_30 <- heart_liver_30[which((as.integer(heart_liver_30[,9]) > 0) == TRUE),]
check_40 <- heart_liver_40[which((as.integer(heart_liver_40[,9]) > 0) == TRUE),]
check_50 <- heart_liver_50[which((as.integer(heart_liver_50[,9]) > 0) == TRUE),]
check_60 <- heart_liver_60[which((as.integer(heart_liver_60[,9]) > 0) == TRUE),]
check_70 <- heart_liver_70[which((as.integer(heart_liver_70[,9]) > 0) == TRUE),]
check_75 <- heart_liver_75[which((as.integer(heart_liver_75[,9]) > 0) == TRUE),]
check_80 <- heart_liver_80[which((as.integer(heart_liver_80[,9]) > 0) == TRUE),]
check_90 <- heart_liver_90[which((as.integer(heart_liver_90[,9]) > 0) == TRUE),]
check_100 <- heart_liver_100[which((as.integer(heart_liver_100[,9]) > 0) == TRUE),]
summary(as.integer(check_1[,9])) Min. 1st Qu. Median Mean 3rd Qu. Max.
2.0 209.0 404.0 509.5 705.8 3679.0 summary(as.integer(check_10[,9])) Min. 1st Qu. Median Mean 3rd Qu. Max.
12.0 231.0 418.0 526.2 723.2 3679.0 summary(as.integer(check_20[,9])) Min. 1st Qu. Median Mean 3rd Qu. Max.
12.0 264.0 451.0 556.7 761.0 3679.0 summary(as.integer(check_25[,9])) Min. 1st Qu. Median Mean 3rd Qu. Max.
12.0 281.0 473.0 574.0 781.8 3679.0 summary(as.integer(check_30[,9])) Min. 1st Qu. Median Mean 3rd Qu. Max.
12.0 295.5 497.0 590.4 802.0 3679.0 summary(as.integer(check_40[,9])) Min. 1st Qu. Median Mean 3rd Qu. Max.
12.0 321.0 533.0 622.4 835.0 3679.0 summary(as.integer(check_50[,9])) Min. 1st Qu. Median Mean 3rd Qu. Max.
12.0 342.0 564.0 652.9 874.0 3679.0 summary(as.integer(check_60[,9])) Min. 1st Qu. Median Mean 3rd Qu. Max.
12.0 356.0 590.0 680.4 920.5 3679.0 summary(as.integer(check_70[,9])) Min. 1st Qu. Median Mean 3rd Qu. Max.
12.0 350.0 619.5 702.7 958.0 3679.0 summary(as.integer(check_75[,9])) Min. 1st Qu. Median Mean 3rd Qu. Max.
12.0 342.5 621.0 709.8 979.0 3679.0 summary(as.integer(check_80[,9])) Min. 1st Qu. Median Mean 3rd Qu. Max.
12 320 602 701 978 3590 summary(as.integer(check_90[,9])) Min. 1st Qu. Median Mean 3rd Qu. Max.
12.0 242.2 501.0 637.5 922.0 3590.0 summary(as.integer(check_100[,9])) Min. 1st Qu. Median Mean 3rd Qu. Max.
12.0 182.0 378.0 489.2 710.5 2266.0 # Plot the median tDMR length
conserved_tdmr_heart_liver_heart_lung_bp <- c(404, 418, 450.5, 473, 497, 533, 564, 590, 619.5, 621,602,501, 378)
perc_overlap <- c(0.000000001, 0.1, 0.2, 0.25, 0.3, 0.4, 0.5, 0.6, 0.7, 0.75, 0.80, 0.90, 1)
plot(perc_overlap, conserved_tdmr_heart_liver_heart_lung_bp, xlab = "Percentage of overlap (bp)", ylab = "Median length of overlap (bp)", pch = 19, main = "Median length of overlap between rhesus tDMRs (HvLi and HvLu)")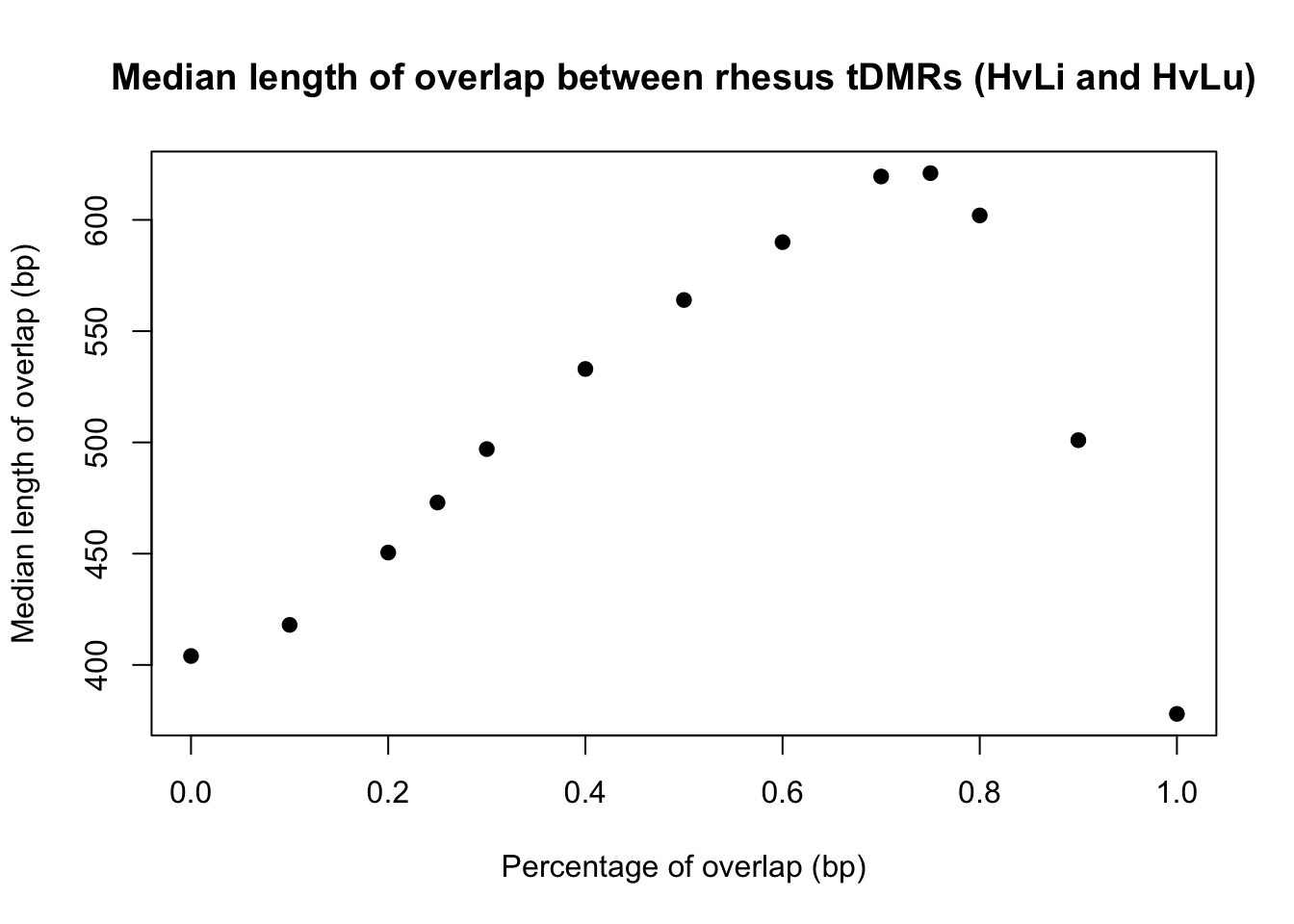
An overlap of 0.70-0.80 maximizes the median length of overlap between tDMRs. This result appears robust to the tissues and species.
Pairwise tDMRs
within_species_tDMR_overlap <- function(filename1, filename2){
# make bed style format: change name from human to rhesus
humans_heart_liver_DMRs_bed <- filename1[,pull_col]
# Get DMRs on the autosomal chromosomes only
humans_heart_liver_DMRs_bed_noX <- humans_heart_liver_DMRs_bed[which(humans_heart_liver_DMRs_bed[,1] != "chrX"),]
humans_heart_liver_DMRs_bed_noX[,1] <- as.character(humans_heart_liver_DMRs_bed_noX[,1])
humans_heart_liver_DMRs_bed_noX[,2] <- as.integer(humans_heart_liver_DMRs_bed_noX[,2])
humans_heart_liver_DMRs_bed_noX[,3] <- as.integer(humans_heart_liver_DMRs_bed_noX[,3])
summary(humans_heart_liver_DMRs_bed_noX[,3] - humans_heart_liver_DMRs_bed_noX[,2])
sort_humans_heart_liver_DMRs_bed <- bedr.sort.region(humans_heart_liver_DMRs_bed_noX, check.chr = FALSE)
# make bed style format
humans_heart_lung_DMRs_bed <- filename2[,pull_col]
humans_heart_lung_DMRs_bed_noX <- humans_heart_lung_DMRs_bed[which(humans_heart_lung_DMRs_bed[,1] != "chrX"),]
humans_heart_lung_DMRs_bed_noX[,1] <- as.character(humans_heart_lung_DMRs_bed_noX[,1])
humans_heart_lung_DMRs_bed_noX[,2] <- as.integer(humans_heart_lung_DMRs_bed_noX[,2])
humans_heart_lung_DMRs_bed_noX[,3] <- as.integer(humans_heart_lung_DMRs_bed_noX[,3])
sort_humans_heart_lung_DMRs_bed <- bedr.sort.region(humans_heart_lung_DMRs_bed_noX, check.chr = FALSE)
summary(humans_heart_lung_DMRs_bed_noX[,3] - humans_heart_lung_DMRs_bed_noX[,2])
heart_lung <- bedr(input = list(a = sort_humans_heart_liver_DMRs_bed, b = sort_humans_heart_lung_DMRs_bed), method = "intersect", params = "-wao -f 0.75 -F 0.75", check.chr = FALSE)
summary(heart_lung[,3] - heart_lung[,2])
summary(as.integer(heart_lung[,9]))
summary(as.integer(heart_lung[,9]) > 0)
lung_heart <- bedr(input = list(a = sort_humans_heart_lung_DMRs_bed, b = sort_humans_heart_liver_DMRs_bed), method = "intersect", params = "-wao -f 0.75 -F 0.75", check.chr = FALSE)
summary(as.integer(lung_heart[,9]))
summary(as.integer(lung_heart[,9]) > 0)
# Make a Venn diagram based on the results
stats_filename1 <- as.array(summary(as.integer(heart_lung[,9]) > 0))
stats_filename2 <- as.array(summary(as.integer(lung_heart[,9]) > 0))
stats_filename12 <- as.data.frame(cbind(stats_filename1[2], stats_filename1[3], stats_filename1[4], stats_filename2[2], stats_filename2[3], stats_filename2[4]))
#stats_filename12[,1] <- as.integer(stats_filename12[,1])
#stats_filename12[,2] <- as.integer(stats_filename12[,2])
#stats_filename12[,3] <- as.integer(stats_filename12[,3])
#stats_filename12[,4] <- as.integer(stats_filename12[,4])
#stats_filename12[,5] <- as.integer(stats_filename12[,5])
#stats_filename12[,6] <- as.integer(stats_filename12[,6])
return(stats_filename12)
}Heart versus liver and heart versus lung
within_species_tDMR_overlap(humans_heart_liver_DMRs, humans_heart_lung_DMRs)SORTING
VALIDATE REGIONS
* Checking input type... PASS
Input is in bed format
* Check if index is a string... PASS
* Check index pattern... PASS
* Check for missing values... PASS
* Check for larger start position... PASS.
* Check if zero based... PASS
* Overlapping regions can cause unexpected results.
SORTING
VALIDATE REGIONS
* Checking input type... PASS
Input is in bed format
* Check if index is a string... PASS
* Check index pattern... PASS
* Check for missing values... PASS
* Check for larger start position... PASS.
* Check if zero based... PASS
* Overlapping regions can cause unexpected results.
* Processing input (1): a
CONVERT TO BED
* Checking input type... PASS
Input is in bed format
VALIDATE REGIONS
* Check if index is a string... PASS
* Check index pattern... PASS
* Check for missing values... PASS
* Check for larger start position... PASS.
* Check if zero based... PASS
* Checking sort order... PASS
* Checking for overlapping 'contiguous' regions... PASS
* Processing input (2): b
CONVERT TO BED
* Checking input type... PASS
Input is in bed format
VALIDATE REGIONS
* Check if index is a string... PASS
* Check index pattern... PASS
* Check for missing values... PASS
* Check for larger start position... PASS.
* Check if zero based... PASS
* Checking sort order... PASS
* Checking for overlapping 'contiguous' regions... PASS
bedtools intersect -a /var/folders/rf/qrcw6ncj05z1pc_pq9xzw3540000gn/T//Rtmph6wW6Y/a_c3e34dd967b6.bed -b /var/folders/rf/qrcw6ncj05z1pc_pq9xzw3540000gn/T//Rtmph6wW6Y/b_c3e32ce3f501.bed -wao -f 0.75 -F 0.75
* Processing input (1): a
CONVERT TO BED
* Checking input type... PASS
Input is in bed format
VALIDATE REGIONS
* Check if index is a string... PASS
* Check index pattern... PASS
* Check for missing values... PASS
* Check for larger start position... PASS.
* Check if zero based... PASS
* Checking sort order... PASS
* Checking for overlapping 'contiguous' regions... PASS
* Processing input (2): b
CONVERT TO BED
* Checking input type... PASS
Input is in bed format
VALIDATE REGIONS
* Check if index is a string... PASS
* Check index pattern... PASS
* Check for missing values... PASS
* Check for larger start position... PASS.
* Check if zero based... PASS
* Checking sort order... PASS
* Checking for overlapping 'contiguous' regions... PASS
bedtools intersect -a /var/folders/rf/qrcw6ncj05z1pc_pq9xzw3540000gn/T//Rtmph6wW6Y/a_c3e35b79bc20.bed -b /var/folders/rf/qrcw6ncj05z1pc_pq9xzw3540000gn/T//Rtmph6wW6Y/b_c3e3149487ca.bed -wao -f 0.75 -F 0.75 V1 V2 V3 V4 V5 V6
FALSE 20256 2305 <NA> 11903 2305 <NA>grid.newpage()
draw.pairwise.venn(area1 = (20256+2305), area2 = (11903+2305), cross.area = 2305, category = c("Human Heart v. Liver", "Heart v. Lung"))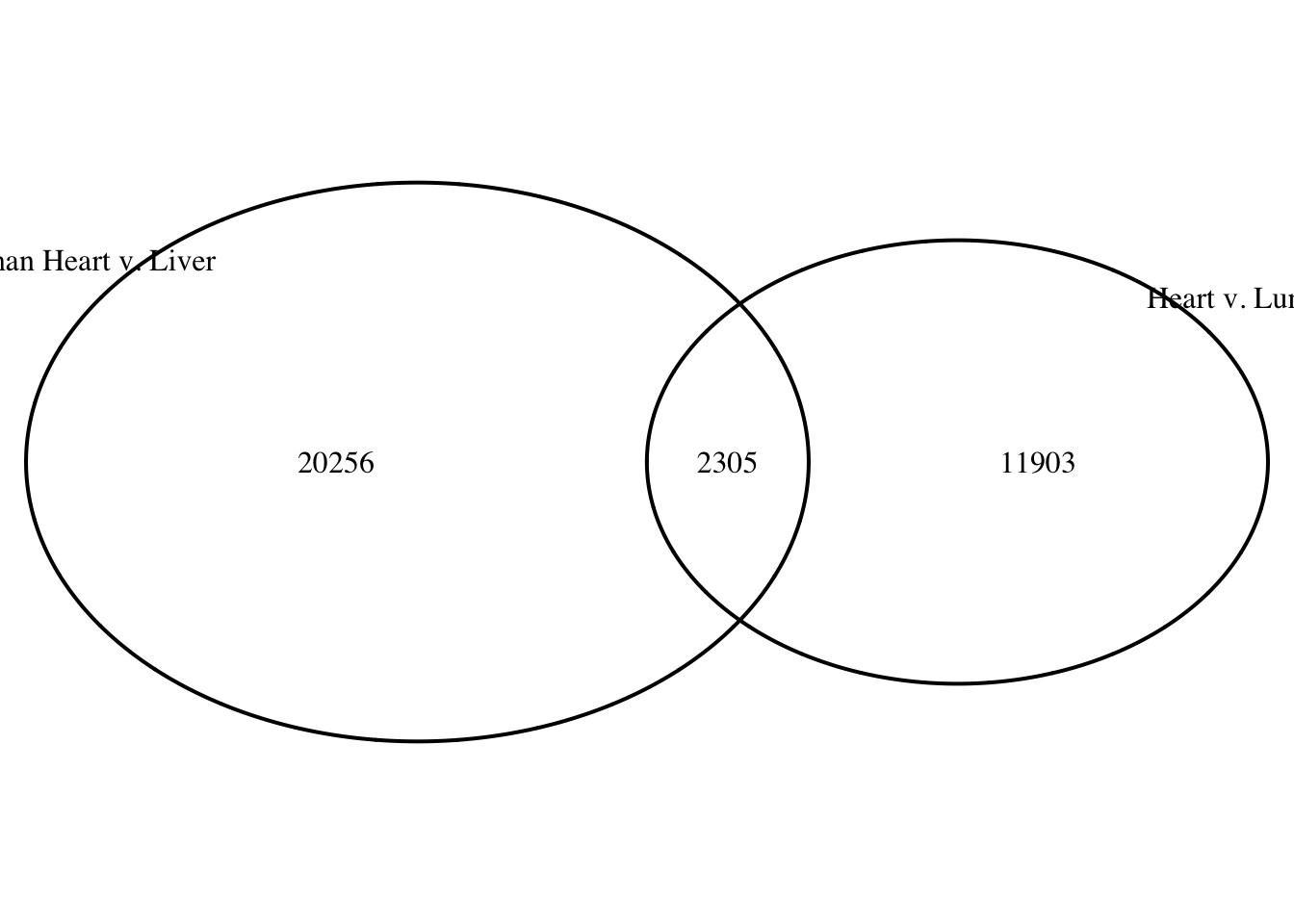
(polygon[GRID.polygon.38], polygon[GRID.polygon.39], polygon[GRID.polygon.40], polygon[GRID.polygon.41], text[GRID.text.42], text[GRID.text.43], text[GRID.text.44], text[GRID.text.45], text[GRID.text.46]) within_species_tDMR_overlap(chimps_heart_liver_DMRs, chimps_heart_lung_DMRs)SORTING
VALIDATE REGIONS
* Checking input type... PASS
Input is in bed format
* Check if index is a string... PASS
* Check index pattern... PASS
* Check for missing values... PASS
* Check for larger start position... PASS.
* Check if zero based... PASS
* Overlapping regions can cause unexpected results.
SORTING
VALIDATE REGIONS
* Checking input type... PASS
Input is in bed format
* Check if index is a string... PASS
* Check index pattern... PASS
* Check for missing values... PASS
* Check for larger start position... PASS.
* Check if zero based... PASS
* Overlapping regions can cause unexpected results.
* Processing input (1): a
CONVERT TO BED
* Checking input type... PASS
Input is in bed format
VALIDATE REGIONS
* Check if index is a string... PASS
* Check index pattern... PASS
* Check for missing values... PASS
* Check for larger start position... PASS.
* Check if zero based... PASS
* Checking sort order... PASS
* Checking for overlapping 'contiguous' regions... PASS
* Processing input (2): b
CONVERT TO BED
* Checking input type... PASS
Input is in bed format
VALIDATE REGIONS
* Check if index is a string... PASS
* Check index pattern... PASS
* Check for missing values... PASS
* Check for larger start position... PASS.
* Check if zero based... PASS
* Checking sort order... PASS
* Checking for overlapping 'contiguous' regions... PASS
bedtools intersect -a /var/folders/rf/qrcw6ncj05z1pc_pq9xzw3540000gn/T//Rtmph6wW6Y/a_c3e3511f4114.bed -b /var/folders/rf/qrcw6ncj05z1pc_pq9xzw3540000gn/T//Rtmph6wW6Y/b_c3e35cf72fe.bed -wao -f 0.75 -F 0.75
* Processing input (1): a
CONVERT TO BED
* Checking input type... PASS
Input is in bed format
VALIDATE REGIONS
* Check if index is a string... PASS
* Check index pattern... PASS
* Check for missing values... PASS
* Check for larger start position... PASS.
* Check if zero based... PASS
* Checking sort order... PASS
* Checking for overlapping 'contiguous' regions... PASS
* Processing input (2): b
CONVERT TO BED
* Checking input type... PASS
Input is in bed format
VALIDATE REGIONS
* Check if index is a string... PASS
* Check index pattern... PASS
* Check for missing values... PASS
* Check for larger start position... PASS.
* Check if zero based... PASS
* Checking sort order... PASS
* Checking for overlapping 'contiguous' regions... PASS
bedtools intersect -a /var/folders/rf/qrcw6ncj05z1pc_pq9xzw3540000gn/T//Rtmph6wW6Y/a_c3e346110dfb.bed -b /var/folders/rf/qrcw6ncj05z1pc_pq9xzw3540000gn/T//Rtmph6wW6Y/b_c3e32d37d33f.bed -wao -f 0.75 -F 0.75 V1 V2 V3 V4 V5 V6
FALSE 27267 1502 <NA> 7024 1502 <NA>grid.newpage()
draw.pairwise.venn(area1 = (27267+1502), area2 = (7024+1502), cross.area = 1502, category = c("Chimp Heart v. Liver", "Heart v. Lung"))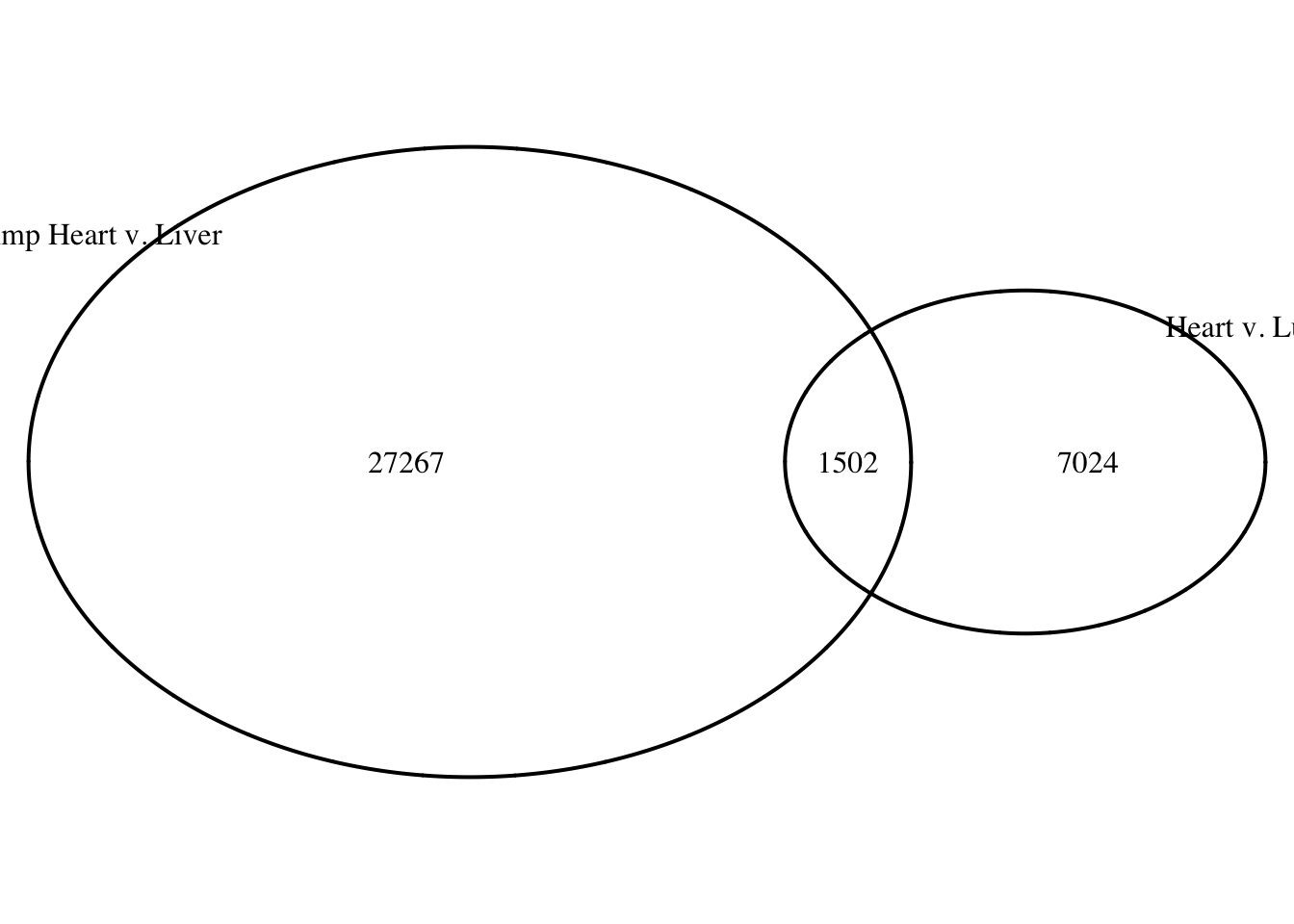
(polygon[GRID.polygon.47], polygon[GRID.polygon.48], polygon[GRID.polygon.49], polygon[GRID.polygon.50], text[GRID.text.51], text[GRID.text.52], text[GRID.text.53], text[GRID.text.54], text[GRID.text.55]) within_species_tDMR_overlap(rhesus_heart_liver_DMRs, rhesus_heart_lung_DMRs)SORTING
VALIDATE REGIONS
* Checking input type... PASS
Input is in bed format
* Check if index is a string... PASS
* Check index pattern... PASS
* Check for missing values... PASS
* Check for larger start position... PASS.
* Check if zero based... PASS
* Overlapping regions can cause unexpected results.
SORTING
VALIDATE REGIONS
* Checking input type... PASS
Input is in bed format
* Check if index is a string... PASS
* Check index pattern... PASS
* Check for missing values... PASS
* Check for larger start position... PASS.
* Check if zero based... PASS
* Overlapping regions can cause unexpected results.
* Processing input (1): a
CONVERT TO BED
* Checking input type... PASS
Input is in bed format
VALIDATE REGIONS
* Check if index is a string... PASS
* Check index pattern... PASS
* Check for missing values... PASS
* Check for larger start position... PASS.
* Check if zero based... PASS
* Checking sort order... PASS
* Checking for overlapping 'contiguous' regions... PASS
* Processing input (2): b
CONVERT TO BED
* Checking input type... PASS
Input is in bed format
VALIDATE REGIONS
* Check if index is a string... PASS
* Check index pattern... PASS
* Check for missing values... PASS
* Check for larger start position... PASS.
* Check if zero based... PASS
* Checking sort order... PASS
* Checking for overlapping 'contiguous' regions... PASS
bedtools intersect -a /var/folders/rf/qrcw6ncj05z1pc_pq9xzw3540000gn/T//Rtmph6wW6Y/a_c3e32181876a.bed -b /var/folders/rf/qrcw6ncj05z1pc_pq9xzw3540000gn/T//Rtmph6wW6Y/b_c3e313de3ad8.bed -wao -f 0.75 -F 0.75
* Processing input (1): a
CONVERT TO BED
* Checking input type... PASS
Input is in bed format
VALIDATE REGIONS
* Check if index is a string... PASS
* Check index pattern... PASS
* Check for missing values... PASS
* Check for larger start position... PASS.
* Check if zero based... PASS
* Checking sort order... PASS
* Checking for overlapping 'contiguous' regions... PASS
* Processing input (2): b
CONVERT TO BED
* Checking input type... PASS
Input is in bed format
VALIDATE REGIONS
* Check if index is a string... PASS
* Check index pattern... PASS
* Check for missing values... PASS
* Check for larger start position... PASS.
* Check if zero based... PASS
* Checking sort order... PASS
* Checking for overlapping 'contiguous' regions... PASS
bedtools intersect -a /var/folders/rf/qrcw6ncj05z1pc_pq9xzw3540000gn/T//Rtmph6wW6Y/a_c3e346542fc6.bed -b /var/folders/rf/qrcw6ncj05z1pc_pq9xzw3540000gn/T//Rtmph6wW6Y/b_c3e3492395ff.bed -wao -f 0.75 -F 0.75 V1 V2 V3 V4 V5 V6
FALSE 39998 1288 <NA> 5739 1288 <NA>grid.newpage()
draw.pairwise.venn(area1 = (39998+1288), area2 = (5739+1288), cross.area = 1288, category = c("Chimp Heart v. Liver", "Heart v. Lung"))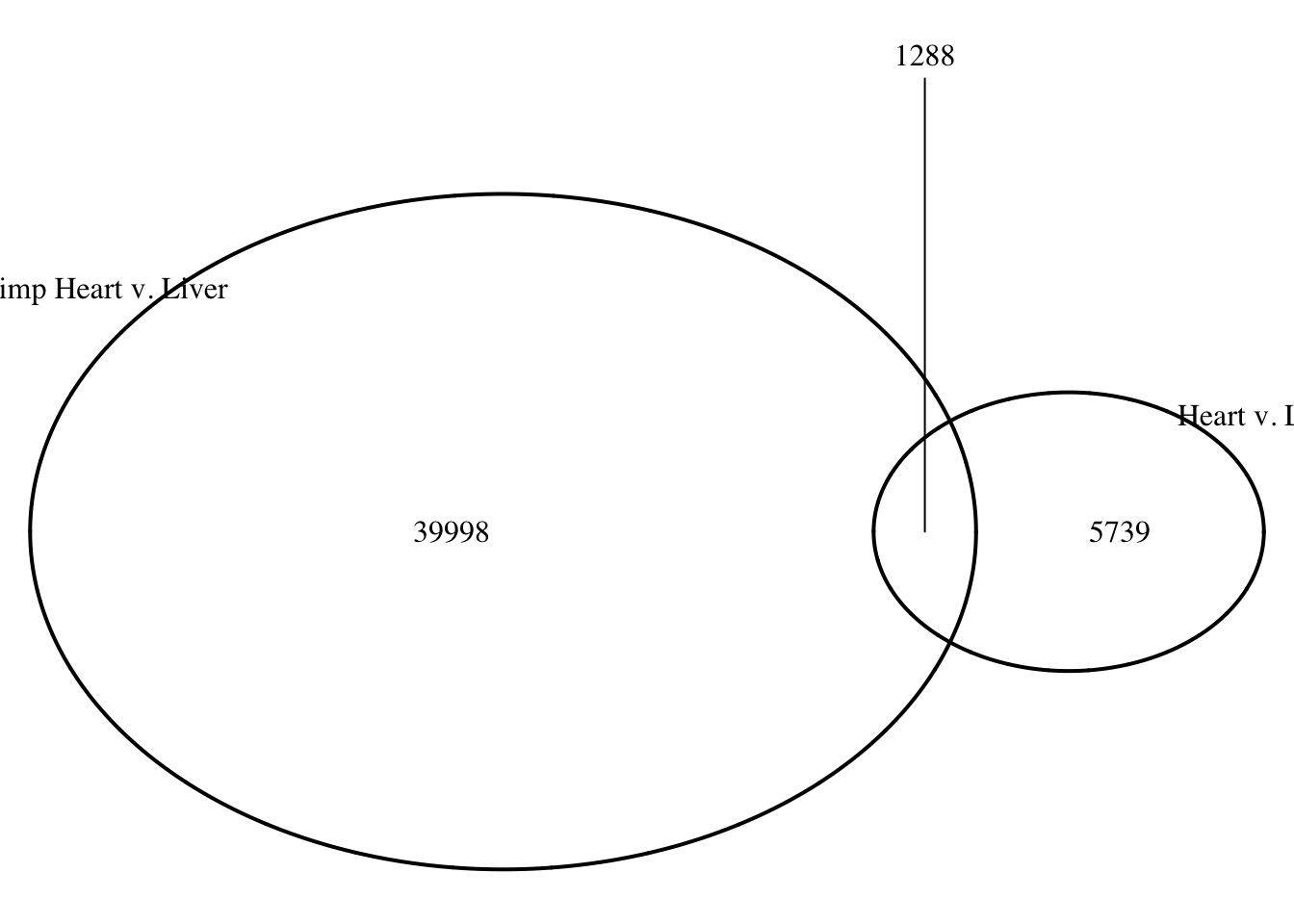
(polygon[GRID.polygon.56], polygon[GRID.polygon.57], polygon[GRID.polygon.58], polygon[GRID.polygon.59], text[GRID.text.60], text[GRID.text.61], text[GRID.text.62], lines[GRID.lines.63], text[GRID.text.64], text[GRID.text.65]) Which overlap promoters?
# Import data about orthologous promoters
boundaries_within_250_humans <- read.delim("~/Desktop/Reg_Evo_Primates/data/250_complete_interval_humans.bed")
boundaries_within_250_humans[,1] <- as.character(boundaries_within_250_humans[,1])
# How many tDMRs cover various percentages of the orthologous promoters
sort_boundaries_within_250_humans <- bedr.sort.region(boundaries_within_250_humans, check.chr = FALSE)SORTING
VALIDATE REGIONS
* Checking input type... PASS
Input seems to be in bed format but chr/start/end column names are missing
* Check if index is a string... PASS
* Check index pattern... PASS
* Check for missing values... PASS
* Check for larger start position... PASS.
* Check if zero based... PASSsort_boundaries_within_250_humans <- bedr.merge.region(boundaries_within_250_humans, check.chr = FALSE)MERGING
VALIDATE REGIONS
* Checking input type... PASS
Input seems to be in bed format but chr/start/end column names are missing
* Check if index is a string... PASS
* Check index pattern... PASS
* Check for missing values... PASS
* Check for larger start position... PASS.
* Check if zero based... PASS
* Processing input (1): i
CONVERT TO BED
* Checking input type... PASS
Input seems to be in bed format but chr/start/end column names are missing
bedtools merge -i /var/folders/rf/qrcw6ncj05z1pc_pq9xzw3540000gn/T//Rtmph6wW6Y/i_c3e31fb52313.bed -d 0 -c 4 -o collapse
* Collapsing 4149 --> 3939 regions... NOTE# Various percentages
heart_liver_promoter_10 <- bedr(input = list(a = boundaries_within_250_humans, b = sort_humans_heart_liver_DMRs_bed), method = "intersect", params = "-wao -f 0.1", check.chr = FALSE) * Processing input (1): a
CONVERT TO BED
* Checking input type... PASS
Input seems to be in bed format but chr/start/end column names are missing
VALIDATE REGIONS
* Check if index is a string... PASS
* Check index pattern... PASS
* Check for missing values... PASS
* Check for larger start position... PASS.
* Check if zero based... PASS
* Checking sort order... PASS
* Checking for overlapping 'contiguous' regions... FAIL
The input for object has overlapping features!
This can cause unexpected results for some set operations.
i.e. x <- bedr.merge.region(x)
* Processing input (2): b
CONVERT TO BED
* Checking input type... PASS
Input is in bed format
VALIDATE REGIONS
* Check if index is a string... PASS
* Check index pattern... PASS
* Check for missing values... PASS
* Check for larger start position... PASS.
* Check if zero based... PASS
* Checking sort order... PASS
* Checking for overlapping 'contiguous' regions... PASS
bedtools intersect -a /var/folders/rf/qrcw6ncj05z1pc_pq9xzw3540000gn/T//Rtmph6wW6Y/a_c3e35ea5d932.bed -b /var/folders/rf/qrcw6ncj05z1pc_pq9xzw3540000gn/T//Rtmph6wW6Y/b_c3e372c4e733.bed -wao -f 0.1summary(heart_liver_promoter_10[,11] > 0) Mode FALSE TRUE
logical 3979 171 heart_liver_promoter_50 <- bedr(input = list(a = boundaries_within_250_humans, b = sort_humans_heart_liver_DMRs_bed), method = "intersect", params = "-wao -f 0.5", check.chr = FALSE) * Processing input (1): a
CONVERT TO BED
* Checking input type... PASS
Input seems to be in bed format but chr/start/end column names are missing
VALIDATE REGIONS
* Check if index is a string... PASS
* Check index pattern... PASS
* Check for missing values... PASS
* Check for larger start position... PASS.
* Check if zero based... PASS
* Checking sort order... PASS
* Checking for overlapping 'contiguous' regions... FAIL
The input for object has overlapping features!
This can cause unexpected results for some set operations.
i.e. x <- bedr.merge.region(x)
* Processing input (2): b
CONVERT TO BED
* Checking input type... PASS
Input is in bed format
VALIDATE REGIONS
* Check if index is a string... PASS
* Check index pattern... PASS
* Check for missing values... PASS
* Check for larger start position... PASS.
* Check if zero based... PASS
* Checking sort order... PASS
* Checking for overlapping 'contiguous' regions... PASS
bedtools intersect -a /var/folders/rf/qrcw6ncj05z1pc_pq9xzw3540000gn/T//Rtmph6wW6Y/a_c3e3506b9c1f.bed -b /var/folders/rf/qrcw6ncj05z1pc_pq9xzw3540000gn/T//Rtmph6wW6Y/b_c3e36f0945d0.bed -wao -f 0.5summary(heart_liver_promoter_50[,11] > 0) Mode FALSE TRUE
logical 4030 119 heart_liver_promoter_100 <- bedr(input = list(a = boundaries_within_250_humans, b = sort_humans_heart_liver_DMRs_bed), method = "intersect", params = "-wao -f 1", check.chr = FALSE) * Processing input (1): a
CONVERT TO BED
* Checking input type... PASS
Input seems to be in bed format but chr/start/end column names are missing
VALIDATE REGIONS
* Check if index is a string... PASS
* Check index pattern... PASS
* Check for missing values... PASS
* Check for larger start position... PASS.
* Check if zero based... PASS
* Checking sort order... PASS
* Checking for overlapping 'contiguous' regions... FAIL
The input for object has overlapping features!
This can cause unexpected results for some set operations.
i.e. x <- bedr.merge.region(x)
* Processing input (2): b
CONVERT TO BED
* Checking input type... PASS
Input is in bed format
VALIDATE REGIONS
* Check if index is a string... PASS
* Check index pattern... PASS
* Check for missing values... PASS
* Check for larger start position... PASS.
* Check if zero based... PASS
* Checking sort order... PASS
* Checking for overlapping 'contiguous' regions... PASS
bedtools intersect -a /var/folders/rf/qrcw6ncj05z1pc_pq9xzw3540000gn/T//Rtmph6wW6Y/a_c3e3155a36df.bed -b /var/folders/rf/qrcw6ncj05z1pc_pq9xzw3540000gn/T//Rtmph6wW6Y/b_c3e31fe1a3fe.bed -wao -f 1summary(heart_liver_promoter_100[,11] > 0) Mode FALSE TRUE
logical 4083 66 # What if we look at 100% coverage in the human hearts and lungs?
heart_lung_promoter_100 <- bedr(input = list(a = boundaries_within_250_humans, b = sort_humans_heart_lung_DMRs_bed), method = "intersect", params = "-wao -f 1", check.chr = FALSE) * Processing input (1): a
CONVERT TO BED
* Checking input type... PASS
Input seems to be in bed format but chr/start/end column names are missing
VALIDATE REGIONS
* Check if index is a string... PASS
* Check index pattern... PASS
* Check for missing values... PASS
* Check for larger start position... PASS.
* Check if zero based... PASS
* Checking sort order... PASS
* Checking for overlapping 'contiguous' regions... FAIL
The input for object has overlapping features!
This can cause unexpected results for some set operations.
i.e. x <- bedr.merge.region(x)
* Processing input (2): b
CONVERT TO BED
* Checking input type... PASS
Input is in bed format
VALIDATE REGIONS
* Check if index is a string... PASS
* Check index pattern... PASS
* Check for missing values... PASS
* Check for larger start position... PASS.
* Check if zero based... PASS
* Checking sort order... PASS
* Checking for overlapping 'contiguous' regions... PASS
bedtools intersect -a /var/folders/rf/qrcw6ncj05z1pc_pq9xzw3540000gn/T//Rtmph6wW6Y/a_c3e3331ba4f2.bed -b /var/folders/rf/qrcw6ncj05z1pc_pq9xzw3540000gn/T//Rtmph6wW6Y/b_c3e334dd7b61.bed -wao -f 1summary(heart_lung_promoter_100[,11] > 0) Mode FALSE TRUE
logical 4144 5