Revisions_H3K27ac
Lauren Blake
June 24, 2019
Open files
Note: due to pairwise ordering from another file (tissue_specific_tDMRs), tDMRs labeled as “hypermethylated” are actually hypomethylated in the lung and kidney.
# Library
library(bedr)## Warning: package 'bedr' was built under R version 3.4.4##
##
## ######################
## #### bedr v1.0.7 ####
## ######################
##
## checking binary availability...
## * Checking path for bedtools... PASS
## /usr/local/bin/bedtools
## * Checking path for bedops... FAIL
## * Checking path for tabix... FAIL
## tests and examples will be skipped on R CMD check if binaries are missinglibrary("topGO")## Loading required package: BiocGenerics## Loading required package: parallel##
## Attaching package: 'BiocGenerics'## The following objects are masked from 'package:parallel':
##
## clusterApply, clusterApplyLB, clusterCall, clusterEvalQ,
## clusterExport, clusterMap, parApply, parCapply, parLapply,
## parLapplyLB, parRapply, parSapply, parSapplyLB## The following objects are masked from 'package:stats':
##
## IQR, mad, sd, var, xtabs## The following objects are masked from 'package:base':
##
## anyDuplicated, append, as.data.frame, cbind, colMeans,
## colnames, colSums, do.call, duplicated, eval, evalq, Filter,
## Find, get, grep, grepl, intersect, is.unsorted, lapply,
## lengths, Map, mapply, match, mget, order, paste, pmax,
## pmax.int, pmin, pmin.int, Position, rank, rbind, Reduce,
## rowMeans, rownames, rowSums, sapply, setdiff, sort, table,
## tapply, union, unique, unsplit, which, which.max, which.min## Loading required package: graph## Loading required package: Biobase## Welcome to Bioconductor
##
## Vignettes contain introductory material; view with
## 'browseVignettes()'. To cite Bioconductor, see
## 'citation("Biobase")', and for packages 'citation("pkgname")'.## Loading required package: GO.db## Loading required package: AnnotationDbi## Loading required package: stats4## Loading required package: IRanges## Loading required package: S4Vectors##
## Attaching package: 'S4Vectors'## The following object is masked from 'package:base':
##
## expand.grid## ## Loading required package: SparseM##
## Attaching package: 'SparseM'## The following object is masked from 'package:base':
##
## backsolve##
## groupGOTerms: GOBPTerm, GOMFTerm, GOCCTerm environments built.##
## Attaching package: 'topGO'## The following object is masked from 'package:IRanges':
##
## members#library("biomaRt")
library("clusterProfiler")## Loading required package: DOSE## DOSE v3.4.0 For help: https://guangchuangyu.github.io/DOSE
##
## If you use DOSE in published research, please cite:
## Guangchuang Yu, Li-Gen Wang, Guang-Rong Yan, Qing-Yu He. DOSE: an R/Bioconductor package for Disease Ontology Semantic and Enrichment analysis. Bioinformatics 2015, 31(4):608-609## clusterProfiler v3.6.0 For help: https://guangchuangyu.github.io/clusterProfiler
##
## If you use clusterProfiler in published research, please cite:
## Guangchuang Yu., Li-Gen Wang, Yanyan Han, Qing-Yu He. clusterProfiler: an R package for comparing biological themes among gene clusters. OMICS: A Journal of Integrative Biology. 2012, 16(5):284-287.library("org.Hs.eg.db")## human_chimp_heart_specific_25_conserved_H3K27_HEART <- read.delim("~/Desktop/Regulatory_Evol/ashlar-trial/data/overlap_reg/overlap_25/human_chimp_heart_specific_25_conserved_H3K27_HEART.bed", header=FALSE, stringsAsFactors = FALSE)
summary(human_chimp_heart_specific_25_conserved_H3K27_HEART$V4)## Length Class Mode
## 455 character character413/(413+42)## [1] 0.9076923human_chimp_heart_specific_25_conserved_H3K27_KIDNEY <- read.delim("~/Desktop/Regulatory_Evol/ashlar-trial/data/overlap_reg/overlap_25/human_chimp_kidney_specific_25_conserved_H3K27_KIDNEY.bed", header=FALSE, stringsAsFactors = FALSE)
summary(human_chimp_heart_specific_25_conserved_H3K27_KIDNEY$V4)## Length Class Mode
## 897 character character858/(858+39)## [1] 0.9565217human_chimp_heart_specific_25_conserved_H3K27_LIVER <- read.delim("~/Desktop/Regulatory_Evol/ashlar-trial/data/overlap_reg/overlap_25/human_chimp_liver_specific_25_conserved_H3K27_LIVER.bed", header=FALSE, stringsAsFactors = FALSE)
summary(human_chimp_heart_specific_25_conserved_H3K27_LIVER$V4)## Length Class Mode
## 1333 character character1081/(1081+252)## [1] 0.8109527human_chimp_heart_specific_25_conserved_H3K27_LUNG <- read.delim("~/Desktop/Regulatory_Evol/ashlar-trial/data/overlap_reg/overlap_25/human_chimp_lung_specific_25_conserved_H3K27_LUNG.bed", header=FALSE, stringsAsFactors = FALSE)
summary(human_chimp_heart_specific_25_conserved_H3K27_LUNG$V4)## Length Class Mode
## 51 character character30/(30+21)## [1] 0.5882353refGene_hg19_TSS <- read.delim("~/Desktop/Regulatory_Evol/ashlar-trial/data/overlap_reg/overlap_25/refGene_hg19_TSS.R", header=FALSE, stringsAsFactors = FALSE)
# Adjust so that bedtools will accept it
refGene_hg19_TSS[,3] <- refGene_hg19_TSS[,3] + 1
# Now sort
human_chimp_heart_cons <- bedr.sort.region(human_chimp_heart_specific_25_conserved_H3K27_HEART[,1:3])## SORTING
## VALIDATE REGIONS
## * Checking input type... PASS
## Input seems to be in bed format but chr/start/end column names are missing
## * Check if index is a string... PASS
## * Check index pattern... PASS
## * Check for missing values... PASS
## * Check for larger start position... PASS.
## * Check if zero based... PASS
## * Overlapping regions can cause unexpected results.refGene_hg19_TSS <- bedr.sort.region(refGene_hg19_TSS)## SORTING
## VALIDATE REGIONS
## * Checking input type... PASS
## Input seems to be in bed format but chr/start/end column names are missing
## * Check if index is a string... PASS
## * Check index pattern... PASS
## * Check for missing values... PASS
## * Check for larger start position... PASS.
## * Check if zero based... PASSclosest_heart <- bedr(input = list(a = human_chimp_heart_specific_25_conserved_H3K27_HEART[,1:3], b = refGene_hg19_TSS), method = "closest", check.chr = FALSE)## * Processing input (1): a
## CONVERT TO BED
## * Checking input type... PASS
## Input seems to be in bed format but chr/start/end column names are missing
## VALIDATE REGIONS
## * Check if index is a string... PASS
## * Check index pattern... PASS
## * Check for missing values... PASS
## * Check for larger start position... PASS.
## * Check if zero based... PASS
## * Checking sort order... PASS
## * Checking for overlapping 'contiguous' regions... PASS
## * Processing input (2): b
## CONVERT TO BED
## * Checking input type... PASS
## Input seems to be in bed format but chr/start/end column names are missing
## VALIDATE REGIONS
## * Check if index is a string... PASS
## * Check index pattern... PASS
## * Check for missing values... PASS
## * Check for larger start position... PASS.
## * Check if zero based... PASS
## * Checking sort order... FAIL
## The input for object is not *lexographically* ordered!
## This can cause unexpected results for some set operations.
## try: x <- bedr.sort.region(x)
## * Checking for overlapping 'contiguous' regions... FAIL
## The input for object has overlapping features!
## This can cause unexpected results for some set operations.
## i.e. x <- bedr.merge.region(x)
## bedtools closest -a /var/folders/rf/qrcw6ncj05z1pc_pq9xzw3540000gn/T//RtmpXK3I56/a_bc557225e1f3.bed -b /var/folders/rf/qrcw6ncj05z1pc_pq9xzw3540000gn/T//RtmpXK3I56/b_bc5530e100bf.bed# Sort based on upsteam/downstream
closest_heart_upstream <- closest_heart[which(closest_heart$V9 == "+"),]
closest_heart_downstream <- closest_heart[which(closest_heart$V9 == "-"),]
distance_heart_upstream <- as.numeric(closest_heart_upstream$V3) - as.numeric(closest_heart_upstream$V5)
distance_heart_downstream <- as.numeric(closest_heart_downstream$V5) - as.numeric(closest_heart_downstream$V3)
length(which(distance_heart_upstream > 0))/length(distance_heart_upstream)## [1] 0.6037152length(which(distance_heart_downstream > 0))/length(distance_heart_downstream)## [1] 0.6140351summary(distance_heart_downstream)## Min. 1st Qu. Median Mean 3rd Qu. Max.
## -751049 -12592 3472 15480 50862 386993Kidney
# Now sort
human_chimp_heart_cons <- bedr.sort.region(human_chimp_heart_specific_25_conserved_H3K27_KIDNEY[,1:3])## SORTING
## VALIDATE REGIONS
## * Checking input type... PASS
## Input seems to be in bed format but chr/start/end column names are missing
## * Check if index is a string... PASS
## * Check index pattern... PASS
## * Check for missing values... PASS
## * Check for larger start position... PASS.
## * Check if zero based... PASSrefGene_hg19_TSS <- bedr.sort.region(refGene_hg19_TSS)## SORTING
## VALIDATE REGIONS
## * Check if index is a string... PASS
## * Check index pattern... PASS
## * Check for missing values... PASS
## * Check for larger start position... PASS.
## * Check if zero based... PASSclosest_heart <- bedr(input = list(a = human_chimp_heart_cons, b = refGene_hg19_TSS), method = "closest", check.chr = FALSE)## * Processing input (1): a
## CONVERT TO BED
## * Checking input type... PASS
## Input seems to be in bed format but chr/start/end column names are missing
## VALIDATE REGIONS
## * Check if index is a string... PASS
## * Check index pattern... PASS
## * Check for missing values... PASS
## * Check for larger start position... PASS.
## * Check if zero based... PASS
## * Checking sort order... PASS
## * Checking for overlapping 'contiguous' regions... FAIL
## The input for object has overlapping features!
## This can cause unexpected results for some set operations.
## i.e. x <- bedr.merge.region(x)
## * Processing input (2): b
## CONVERT TO BED
## * Checking input type... PASS
## Input seems to be in bed format but chr/start/end column names are missing
## VALIDATE REGIONS
## * Check if index is a string... PASS
## * Check index pattern... PASS
## * Check for missing values... PASS
## * Check for larger start position... PASS.
## * Check if zero based... PASS
## * Checking sort order... FAIL
## The input for object is not *lexographically* ordered!
## This can cause unexpected results for some set operations.
## try: x <- bedr.sort.region(x)
## * Checking for overlapping 'contiguous' regions... FAIL
## The input for object has overlapping features!
## This can cause unexpected results for some set operations.
## i.e. x <- bedr.merge.region(x)
## bedtools closest -a /var/folders/rf/qrcw6ncj05z1pc_pq9xzw3540000gn/T//RtmpXK3I56/a_bc553d991179.bed -b /var/folders/rf/qrcw6ncj05z1pc_pq9xzw3540000gn/T//RtmpXK3I56/b_bc551a874cb1.bed# Sort based on upsteam/downstream
closest_heart_upstream <- closest_heart[which(closest_heart$V9 == "+"),]
closest_heart_downstream <- closest_heart[which(closest_heart$V9 == "-"),]
distance_heart_upstream <- as.numeric(closest_heart_upstream$V3) - as.numeric(closest_heart_upstream$V5)
distance_heart_downstream <- as.numeric(closest_heart_downstream$V5) - as.numeric(closest_heart_downstream$V3)
length(which(distance_heart_upstream > 0))/length(distance_heart_upstream)## [1] 0.5685358length(which(distance_heart_downstream > 0))/length(distance_heart_downstream)## [1] 0.5628227Liver
# Now sort
human_chimp_heart_cons <- bedr.sort.region(human_chimp_heart_specific_25_conserved_H3K27_LIVER[,1:3])## SORTING
## VALIDATE REGIONS
## * Checking input type... PASS
## Input seems to be in bed format but chr/start/end column names are missing
## * Check if index is a string... PASS
## * Check index pattern... PASS
## * Check for missing values... PASS
## * Check for larger start position... PASS.
## * Check if zero based... PASSrefGene_hg19_TSS <- bedr.sort.region(refGene_hg19_TSS)## SORTING
## VALIDATE REGIONS
## * Check if index is a string... PASS
## * Check index pattern... PASS
## * Check for missing values... PASS
## * Check for larger start position... PASS.
## * Check if zero based... PASSclosest_heart <- bedr(input = list(a = human_chimp_heart_cons, b = refGene_hg19_TSS), method = "closest", check.chr = FALSE)## * Processing input (1): a
## CONVERT TO BED
## * Checking input type... PASS
## Input seems to be in bed format but chr/start/end column names are missing
## VALIDATE REGIONS
## * Check if index is a string... PASS
## * Check index pattern... PASS
## * Check for missing values... PASS
## * Check for larger start position... PASS.
## * Check if zero based... PASS
## * Checking sort order... PASS
## * Checking for overlapping 'contiguous' regions... FAIL
## The input for object has overlapping features!
## This can cause unexpected results for some set operations.
## i.e. x <- bedr.merge.region(x)
## * Processing input (2): b
## CONVERT TO BED
## * Checking input type... PASS
## Input seems to be in bed format but chr/start/end column names are missing
## VALIDATE REGIONS
## * Check if index is a string... PASS
## * Check index pattern... PASS
## * Check for missing values... PASS
## * Check for larger start position... PASS.
## * Check if zero based... PASS
## * Checking sort order... FAIL
## The input for object is not *lexographically* ordered!
## This can cause unexpected results for some set operations.
## try: x <- bedr.sort.region(x)
## * Checking for overlapping 'contiguous' regions... FAIL
## The input for object has overlapping features!
## This can cause unexpected results for some set operations.
## i.e. x <- bedr.merge.region(x)
## bedtools closest -a /var/folders/rf/qrcw6ncj05z1pc_pq9xzw3540000gn/T//RtmpXK3I56/a_bc551ef7a8be.bed -b /var/folders/rf/qrcw6ncj05z1pc_pq9xzw3540000gn/T//RtmpXK3I56/b_bc551c85df81.bed# Sort based on upsteam/downstream
closest_liver_upstream <- closest_heart[which(closest_heart$V9 == "+"),]
closest_liver_downstream <- closest_heart[which(closest_heart$V9 == "-"),]
distance_liver_upstream <- as.numeric(closest_liver_upstream$V3) - as.numeric(closest_liver_upstream$V5)
distance_liver_downstream <- as.numeric(closest_liver_downstream$V5) - as.numeric(closest_liver_downstream$V3)
length(which(distance_liver_upstream > 0))/length(distance_liver_upstream)## [1] 0.6512043length(which(distance_liver_downstream > 0))/length(distance_liver_downstream)## [1] 0.6230032all_distance <- c(distance_heart_upstream, distance_heart_downstream, distance_liver_upstream, distance_liver_downstream)
summary(all_distance)## Min. 1st Qu. Median Mean 3rd Qu. Max.
## -805181 -7548 1690 13079 31168 999526Lung
# Now sort
human_chimp_heart_cons <- bedr.sort.region(human_chimp_heart_specific_25_conserved_H3K27_LUNG[,1:3])## SORTING
## VALIDATE REGIONS
## * Checking input type... PASS
## Input seems to be in bed format but chr/start/end column names are missing
## * Check if index is a string... PASS
## * Check index pattern... PASS
## * Check for missing values... PASS
## * Check for larger start position... PASS.
## * Check if zero based... PASS
## * Overlapping regions can cause unexpected results.refGene_hg19_TSS <- bedr.sort.region(refGene_hg19_TSS)## SORTING
## VALIDATE REGIONS
## * Check if index is a string... PASS
## * Check index pattern... PASS
## * Check for missing values... PASS
## * Check for larger start position... PASS.
## * Check if zero based... PASSclosest_heart <- bedr(input = list(a = human_chimp_heart_cons, b = refGene_hg19_TSS), method = "closest", check.chr = FALSE)## * Processing input (1): a
## CONVERT TO BED
## * Checking input type... PASS
## Input seems to be in bed format but chr/start/end column names are missing
## VALIDATE REGIONS
## * Check if index is a string... PASS
## * Check index pattern... PASS
## * Check for missing values... PASS
## * Check for larger start position... PASS.
## * Check if zero based... PASS
## * Checking sort order... PASS
## * Checking for overlapping 'contiguous' regions... PASS
## * Processing input (2): b
## CONVERT TO BED
## * Checking input type... PASS
## Input seems to be in bed format but chr/start/end column names are missing
## VALIDATE REGIONS
## * Check if index is a string... PASS
## * Check index pattern... PASS
## * Check for missing values... PASS
## * Check for larger start position... PASS.
## * Check if zero based... PASS
## * Checking sort order... FAIL
## The input for object is not *lexographically* ordered!
## This can cause unexpected results for some set operations.
## try: x <- bedr.sort.region(x)
## * Checking for overlapping 'contiguous' regions... FAIL
## The input for object has overlapping features!
## This can cause unexpected results for some set operations.
## i.e. x <- bedr.merge.region(x)
## bedtools closest -a /var/folders/rf/qrcw6ncj05z1pc_pq9xzw3540000gn/T//RtmpXK3I56/a_bc55282f46b3.bed -b /var/folders/rf/qrcw6ncj05z1pc_pq9xzw3540000gn/T//RtmpXK3I56/b_bc551be5509c.bed# Sort based on upsteam/downstream
closest_heart_upstream <- closest_heart[which(closest_heart$V9 == "+"),]
closest_heart_downstream <- closest_heart[which(closest_heart$V9 == "-"),]
distance_heart_upstream <- as.numeric(closest_heart_upstream$V3) - as.numeric(closest_heart_upstream$V5)
distance_heart_downstream <- as.numeric(closest_heart_downstream$V5) - as.numeric(closest_heart_downstream$V3)
length(which(distance_heart_upstream > 0))/length(distance_heart_upstream)## [1] 0.673913length(which(distance_heart_downstream > 0))/length(distance_heart_downstream)## [1] 0.5714286Separate into hypermethylated and overlapping H3K27ac
Heart
# Find the heart hypo
heart_hypo <- human_chimp_heart_specific_25_conserved_H3K27_HEART[which(human_chimp_heart_specific_25_conserved_H3K27_LIVER$V4 == "hyper" & human_chimp_heart_specific_25_conserved_H3K27_LIVER$V6 != "-1"),]
heart_hypo <- heart_hypo[complete.cases(heart_hypo), ]
# Find the closest gene
closest_heart <- bedr(input = list(a = heart_hypo[,1:3], b = refGene_hg19_TSS), method = "closest", check.chr = FALSE)## * Processing input (1): a
## CONVERT TO BED
## * Checking input type... PASS
## Input seems to be in bed format but chr/start/end column names are missing
## VALIDATE REGIONS
## * Check if index is a string... PASS
## * Check index pattern... PASS
## * Check for missing values... PASS
## * Check for larger start position... PASS.
## * Check if zero based... PASS
## * Checking sort order... PASS
## * Checking for overlapping 'contiguous' regions... PASS
## * Processing input (2): b
## CONVERT TO BED
## * Checking input type... PASS
## Input seems to be in bed format but chr/start/end column names are missing
## VALIDATE REGIONS
## * Check if index is a string... PASS
## * Check index pattern... PASS
## * Check for missing values... PASS
## * Check for larger start position... PASS.
## * Check if zero based... PASS
## * Checking sort order... FAIL
## The input for object is not *lexographically* ordered!
## This can cause unexpected results for some set operations.
## try: x <- bedr.sort.region(x)
## * Checking for overlapping 'contiguous' regions... FAIL
## The input for object has overlapping features!
## This can cause unexpected results for some set operations.
## i.e. x <- bedr.merge.region(x)
## bedtools closest -a /var/folders/rf/qrcw6ncj05z1pc_pq9xzw3540000gn/T//RtmpXK3I56/a_bc55367de709.bed -b /var/folders/rf/qrcw6ncj05z1pc_pq9xzw3540000gn/T//RtmpXK3I56/b_bc55788e817d.bed# Convert the gene name to ensg
gene_id <- read.table("../../../Reg_Evo_Primates/data/ENSG_GENE_HG19.csv", stringsAsFactors = FALSE, header=TRUE, sep = ",")
comb_kidney <- merge(closest_heart, gene_id, by.x = c("V8"), by.y = c("Gene"))
comb_kidney$ensg## [1] "ENSG00000196839" "ENSG00000163050" "ENSG00000178878"
## [4] "ENSG00000130762" "ENSG00000168646" "ENSG00000110092"
## [7] "ENSG00000106554" "ENSG00000134871" "ENSG00000244274"
## [10] "ENSG00000244274" "ENSG00000135636" "ENSG00000135636"
## [13] "ENSG00000135636" "ENSG00000135636" "ENSG00000135636"
## [16] "ENSG00000135636" "ENSG00000135636" "ENSG00000108001"
## [19] "ENSG00000187773" "ENSG00000139132" "ENSG00000153303"
## [22] "ENSG00000143641" "ENSG00000144596" "ENSG00000115756"
## [25] "ENSG00000197081" "ENSG00000185507" "ENSG00000123104"
## [28] "ENSG00000127528" "ENSG00000106003" "ENSG00000107902"
## [31] "ENSG00000107902" "ENSG00000183060" "ENSG00000068305"
## [34] "ENSG00000068305" "ENSG00000068305" "ENSG00000068305"
## [37] "ENSG00000120254" "ENSG00000120254" "ENSG00000120254"
## [40] "ENSG00000133454" "ENSG00000124785" "ENSG00000162407"
## [43] "ENSG00000107263" "ENSG00000159788" "ENSG00000138835"
## [46] "ENSG00000064932" "ENSG00000104969" "ENSG00000130147"
## [49] "ENSG00000156222" "ENSG00000156222" "ENSG00000156222"
## [52] "ENSG00000156222" "ENSG00000095637" "ENSG00000095637"
## [55] "ENSG00000185594" "ENSG00000065882" "ENSG00000109927"
## [58] "ENSG00000100726" "ENSG00000185361" "ENSG00000198795"
## [61] "ENSG00000198597" "ENSG00000141579" "ENSG00000070476"
## [64] "ENSG00000070476"# Convert the TSS to ensg as well
ref_gene_hg19_ensg <- merge(refGene_hg19_TSS, gene_id, by.x = c("V5"), by.y = c("Gene"))
all_ref_gene_hg19_ensg <- unique(ref_gene_hg19_ensg$ensg)
heart_ref_gene <- all_ref_gene_hg19_ensg %in% comb_kidney$ensg
# Revisions- run GO
# Merge ENSG with true/false
test_gene <- as.numeric(as.vector(heart_ref_gene))
names(test_gene) <- all_ref_gene_hg19_ensg
# Run topGO
go_data <- new("topGOdata",
ontology = "BP",
allGenes = test_gene,
geneSel = function(allScore){
return(allScore > 0)
},
nodeSize = 5,
annotationFun = annFUN.org,
mapping = "org.Hs.eg.db",
ID = "ensembl")##
## Building most specific GOs .....## ( 11450 GO terms found. )##
## Build GO DAG topology ..........## ( 15456 GO terms and 36153 relations. )##
## Annotating nodes ...............## ( 14505 genes annotated to the GO terms. )# Perform enrichment test
go_test <- runTest(go_data, algorithm = "weight01", statistic = "fisher")##
## -- Weight01 Algorithm --
##
## the algorithm is scoring 1568 nontrivial nodes
## parameters:
## test statistic: fisher##
## Level 15: 1 nodes to be scored (0 eliminated genes)##
## Level 14: 11 nodes to be scored (0 eliminated genes)##
## Level 13: 29 nodes to be scored (103 eliminated genes)##
## Level 12: 50 nodes to be scored (298 eliminated genes)##
## Level 11: 72 nodes to be scored (2501 eliminated genes)##
## Level 10: 107 nodes to be scored (3937 eliminated genes)##
## Level 9: 161 nodes to be scored (5541 eliminated genes)##
## Level 8: 201 nodes to be scored (7251 eliminated genes)##
## Level 7: 245 nodes to be scored (8558 eliminated genes)##
## Level 6: 265 nodes to be scored (10615 eliminated genes)##
## Level 5: 217 nodes to be scored (12035 eliminated genes)##
## Level 4: 127 nodes to be scored (13480 eliminated genes)##
## Level 3: 63 nodes to be scored (13985 eliminated genes)##
## Level 2: 18 nodes to be scored (14177 eliminated genes)##
## Level 1: 1 nodes to be scored (14301 eliminated genes)go_table <- GenTable(go_data, weightFisher = go_test,
orderBy = "weightFisher", ranksOf = "weightFisher",
topNodes = sum(score(go_test) < .05))
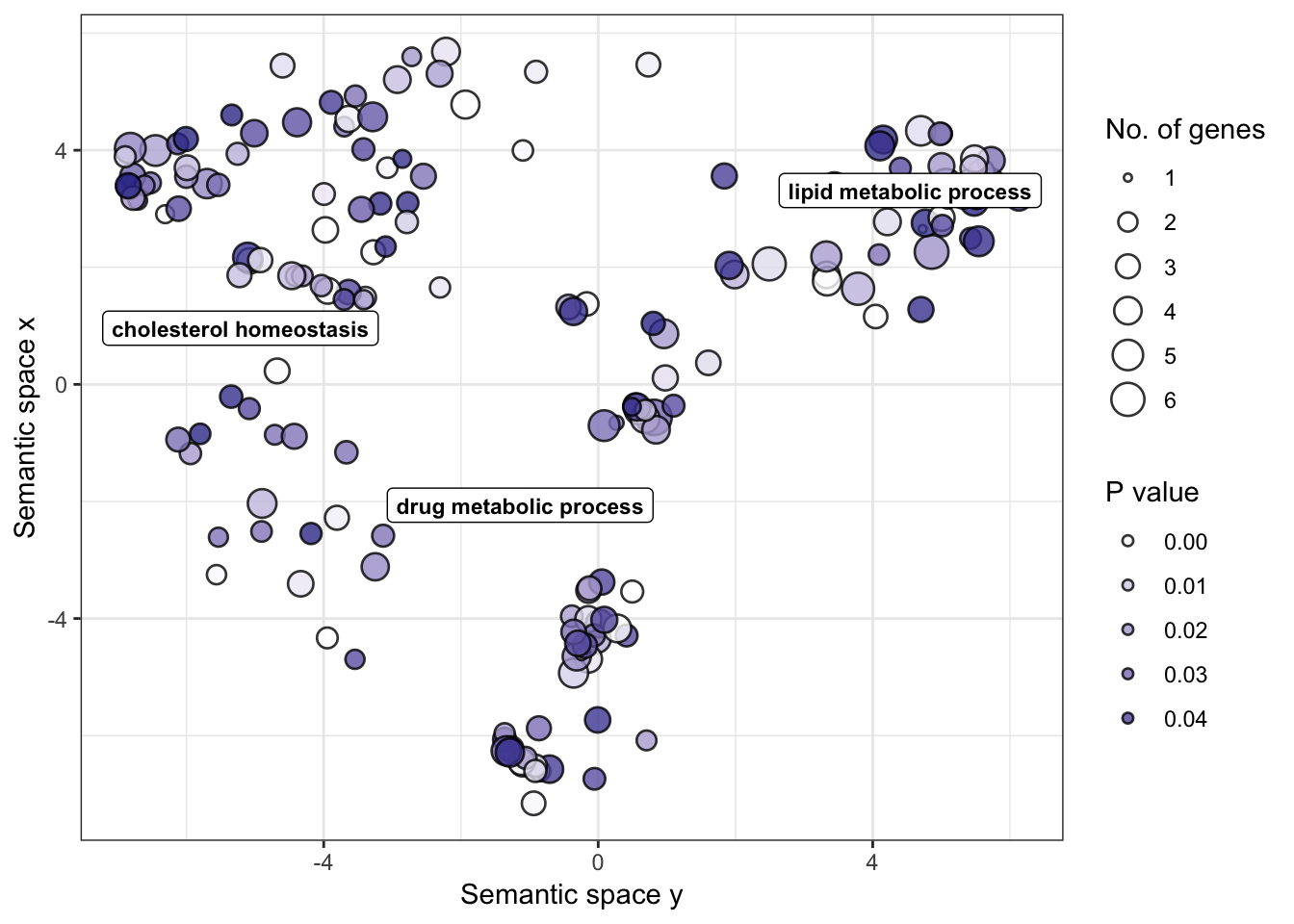
go_table## GO.ID Term Annotated
## 1 GO:0009113 purine nucleobase biosynthetic process 12
## 2 GO:0033197 response to vitamin E 12
## 3 GO:0006351 transcription, DNA-templated 3277
## 4 GO:0032007 negative regulation of TOR signaling 40
## 5 GO:0043547 positive regulation of GTPase activity 354
## 6 GO:0060070 canonical Wnt signaling pathway 274
## 7 GO:1903078 positive regulation of protein localizat... 46
## 8 GO:0071320 cellular response to cAMP 47
## 9 GO:0006501 C-terminal protein lipidation 62
## 10 GO:0007386 compartment pattern specification 5
## 11 GO:0070375 ERK5 cascade 5
## 12 GO:0042321 negative regulation of circadian sleep/w... 5
## 13 GO:0009146 purine nucleoside triphosphate catabolic... 5
## 14 GO:0060509 Type I pneumocyte differentiation 5
## 15 GO:0046060 dATP metabolic process 5
## 16 GO:0009256 10-formyltetrahydrofolate metabolic proc... 5
## 17 GO:0048541 Peyer's patch development 5
## 18 GO:0009168 purine ribonucleoside monophosphate bios... 66
## 19 GO:0006863 purine nucleobase transport 6
## 20 GO:0015942 formate metabolic process 6
## 21 GO:0018242 protein O-linked glycosylation via serin... 6
## 22 GO:0001957 intramembranous ossification 6
## 23 GO:0060368 regulation of Fc receptor mediated stimu... 6
## 24 GO:0070141 response to UV-A 6
## 25 GO:0038203 TORC2 signaling 6
## 26 GO:0033632 regulation of cell-cell adhesion mediate... 6
## 27 GO:0060405 regulation of penile erection 6
## 28 GO:0009204 deoxyribonucleoside triphosphate catabol... 6
## 29 GO:0070244 negative regulation of thymocyte apoptot... 6
## 30 GO:0090630 activation of GTPase activity 76
## 31 GO:0002819 regulation of adaptive immune response 127
## 32 GO:0032261 purine nucleotide salvage 7
## 33 GO:0018243 protein O-linked glycosylation via threo... 7
## 34 GO:0001778 plasma membrane repair 7
## 35 GO:1903071 positive regulation of ER-associated ubi... 7
## 36 GO:0046654 tetrahydrofolate biosynthetic process 7
## 37 GO:0009120 deoxyribonucleoside metabolic process 7
## 38 GO:0019043 establishment of viral latency 7
## 39 GO:0048702 embryonic neurocranium morphogenesis 7
## 40 GO:1901642 nucleoside transmembrane transport 7
## 41 GO:2000109 regulation of macrophage apoptotic proce... 7
## 42 GO:0046130 purine ribonucleoside catabolic process 7
## 43 GO:0033601 positive regulation of mammary gland epi... 7
## 44 GO:0071499 cellular response to laminar fluid shear... 7
## 45 GO:0008277 regulation of G-protein coupled receptor... 120
## 46 GO:1904263 positive regulation of TORC1 signaling 8
## 47 GO:0038180 nerve growth factor signaling pathway 8
## 48 GO:0038063 collagen-activated tyrosine kinase recep... 8
## 49 GO:0060340 positive regulation of type I interferon... 8
## 50 GO:0072530 purine-containing compound transmembrane... 8
## 51 GO:0048703 embryonic viscerocranium morphogenesis 8
## 52 GO:0034127 regulation of MyD88-independent toll-lik... 8
## 53 GO:0008354 germ cell migration 8
## 54 GO:0014807 regulation of somitogenesis 8
## 55 GO:0045188 regulation of circadian sleep/wake cycle... 8
## 56 GO:0048387 negative regulation of retinoic acid rec... 8
## 57 GO:0002313 mature B cell differentiation involved i... 18
## 58 GO:0033089 positive regulation of T cell differenti... 9
## 59 GO:0002315 marginal zone B cell differentiation 9
## 60 GO:0035999 tetrahydrofolate interconversion 9
## 61 GO:0001821 histamine secretion 9
## 62 GO:0003139 secondary heart field specification 9
## 63 GO:0039530 MDA-5 signaling pathway 9
## 64 GO:0002634 regulation of germinal center formation 9
## 65 GO:0070255 regulation of mucus secretion 9
## 66 GO:0001889 liver development 124
## 67 GO:0030282 bone mineralization 93
## 68 GO:0002903 negative regulation of B cell apoptotic ... 10
## 69 GO:0070601 centromeric sister chromatid cohesion 10
## 70 GO:0046085 adenosine metabolic process 10
## 71 GO:0042118 endothelial cell activation 10
## 72 GO:0035414 negative regulation of catenin import in... 10
## 73 GO:0033033 negative regulation of myeloid cell apop... 11
## 74 GO:0001973 adenosine receptor signaling pathway 11
## 75 GO:0003413 chondrocyte differentiation involved in ... 11
## 76 GO:0033327 Leydig cell differentiation 11
## 77 GO:0043249 erythrocyte maturation 11
## 78 GO:1904293 negative regulation of ERAD pathway 12
## 79 GO:0045351 type I interferon biosynthetic process 12
## 80 GO:0055015 ventricular cardiac muscle cell developm... 12
## 81 GO:2000059 negative regulation of protein ubiquitin... 12
## 82 GO:0050862 positive regulation of T cell receptor s... 12
## 83 GO:0048311 mitochondrion distribution 12
## 84 GO:0002281 macrophage activation involved in immune... 12
## 85 GO:0061430 bone trabecula morphogenesis 13
## 86 GO:0043650 dicarboxylic acid biosynthetic process 13
## 87 GO:0071361 cellular response to ethanol 13
## 88 GO:0032486 Rap protein signal transduction 13
## 89 GO:2000107 negative regulation of leukocyte apoptot... 39
## 90 GO:0036066 protein O-linked fucosylation 14
## 91 GO:0045725 positive regulation of glycogen biosynth... 15
## 92 GO:0060749 mammary gland alveolus development 15
## 93 GO:0001829 trophectodermal cell differentiation 15
## 94 GO:0006744 ubiquinone biosynthetic process 15
## 95 GO:0035023 regulation of Rho protein signal transdu... 118
## 96 GO:0051412 response to corticosterone 16
## 97 GO:0051446 positive regulation of meiotic cell cycl... 16
## 98 GO:0072531 pyrimidine-containing compound transmemb... 16
## 99 GO:0055003 cardiac myofibril assembly 16
## 100 GO:0009264 deoxyribonucleotide catabolic process 16
## 101 GO:0000122 negative regulation of transcription fro... 727
## 102 GO:0030111 regulation of Wnt signaling pathway 295
## 103 GO:0046386 deoxyribose phosphate catabolic process 17
## 104 GO:0045655 regulation of monocyte differentiation 17
## 105 GO:0071157 negative regulation of cell cycle arrest 18
## 106 GO:0032026 response to magnesium ion 18
## 107 GO:0046655 folic acid metabolic process 18
## 108 GO:0007063 regulation of sister chromatid cohesion 19
## 109 GO:2000178 negative regulation of neural precursor ... 19
## 110 GO:0006139 nucleobase-containing compound metabolic... 5072
## Significant Expected weightFisher
## 1 2 0.03 0.00046
## 2 2 0.03 0.00046
## 3 14 8.81 0.00408
## 4 2 0.11 0.00515
## 5 6 0.95 0.00540
## 6 4 0.74 0.00656
## 7 2 0.12 0.00677
## 8 2 0.13 0.00706
## 9 2 0.17 0.01203
## 10 1 0.01 0.01337
## 11 1 0.01 0.01337
## 12 1 0.01 0.01337
## 13 1 0.01 0.01337
## 14 1 0.01 0.01337
## 15 1 0.01 0.01337
## 16 1 0.01 0.01337
## 17 1 0.01 0.01337
## 18 2 0.18 0.01356
## 19 1 0.02 0.01603
## 20 1 0.02 0.01603
## 21 1 0.02 0.01603
## 22 1 0.02 0.01603
## 23 1 0.02 0.01603
## 24 1 0.02 0.01603
## 25 1 0.02 0.01603
## 26 1 0.02 0.01603
## 27 1 0.02 0.01603
## 28 1 0.02 0.01603
## 29 1 0.02 0.01603
## 30 2 0.20 0.01771
## 31 2 0.34 0.01835
## 32 1 0.02 0.01867
## 33 1 0.02 0.01867
## 34 1 0.02 0.01867
## 35 1 0.02 0.01867
## 36 1 0.02 0.01867
## 37 1 0.02 0.01867
## 38 1 0.02 0.01867
## 39 1 0.02 0.01867
## 40 1 0.02 0.01867
## 41 1 0.02 0.01867
## 42 1 0.02 0.01867
## 43 1 0.02 0.01867
## 44 1 0.02 0.01867
## 45 3 0.32 0.02091
## 46 1 0.02 0.02131
## 47 1 0.02 0.02131
## 48 1 0.02 0.02131
## 49 1 0.02 0.02131
## 50 1 0.02 0.02131
## 51 1 0.02 0.02131
## 52 1 0.02 0.02131
## 53 1 0.02 0.02131
## 54 1 0.02 0.02131
## 55 1 0.02 0.02131
## 56 1 0.02 0.02131
## 57 2 0.05 0.02335
## 58 1 0.02 0.02395
## 59 1 0.02 0.02395
## 60 1 0.02 0.02395
## 61 1 0.02 0.02395
## 62 1 0.02 0.02395
## 63 1 0.02 0.02395
## 64 1 0.02 0.02395
## 65 1 0.02 0.02395
## 66 3 0.33 0.02423
## 67 2 0.25 0.02584
## 68 1 0.03 0.02657
## 69 1 0.03 0.02657
## 70 1 0.03 0.02657
## 71 1 0.03 0.02657
## 72 1 0.03 0.02657
## 73 1 0.03 0.02919
## 74 1 0.03 0.02919
## 75 1 0.03 0.02919
## 76 1 0.03 0.02919
## 77 1 0.03 0.02919
## 78 1 0.03 0.03180
## 79 1 0.03 0.03180
## 80 1 0.03 0.03180
## 81 1 0.03 0.03180
## 82 1 0.03 0.03180
## 83 1 0.03 0.03180
## 84 1 0.03 0.03180
## 85 1 0.03 0.03441
## 86 1 0.03 0.03441
## 87 1 0.03 0.03441
## 88 1 0.03 0.03441
## 89 2 0.10 0.03614
## 90 1 0.04 0.03701
## 91 1 0.04 0.03960
## 92 1 0.04 0.03960
## 93 1 0.04 0.03960
## 94 1 0.04 0.03960
## 95 2 0.32 0.03999
## 96 1 0.04 0.04218
## 97 1 0.04 0.04218
## 98 1 0.04 0.04218
## 99 1 0.04 0.04218
## 100 1 0.04 0.04218
## 101 5 1.95 0.04391
## 102 4 0.79 0.04460
## 103 1 0.05 0.04476
## 104 1 0.05 0.04476
## 105 1 0.05 0.04733
## 106 1 0.05 0.04733
## 107 1 0.05 0.04733
## 108 1 0.05 0.04990
## 109 1 0.05 0.04990
## 110 19 13.64 0.04991go_table_heart <- as.data.frame(cbind(go_table$GO.ID, go_table$weightFisher))
write.table(go_table_heart, "../data/go_table_heart.txt", quote = FALSE, row.names = FALSE, col.names = FALSE)
# Get names of kidney genes
sig.genes <- sigGenes(go_data)
goresults <- sapply(go_table$GO.ID, function(x)
{
genes<-genesInTerm(go_data, x)
genes[[1]][genes[[1]] %in% sig.genes]
})
# cardiac myofibril assembly, positive regulation of heart rate, regulation of monocyte differentiation
goresults["GO:0055003"] ## $`GO:0055003`
## [1] "ENSG00000068305"goresults["GO:0010460"]## $<NA>
## NULLgoresults["GO:0045655"]## $`GO:0045655`
## [1] "ENSG00000185507"Kidney
# Find the heart hypo
heart_hypo <- human_chimp_heart_specific_25_conserved_H3K27_KIDNEY[which(human_chimp_heart_specific_25_conserved_H3K27_KIDNEY$V4 == "hyper" & human_chimp_heart_specific_25_conserved_H3K27_KIDNEY$V6 != "-1"),]
heart_hypo <- heart_hypo[complete.cases(heart_hypo), ]
# Find the closest gene
closest_heart <- bedr(input = list(a = heart_hypo[,1:3], b = refGene_hg19_TSS), method = "closest", check.chr = FALSE)## * Processing input (1): a
## CONVERT TO BED
## * Checking input type... PASS
## Input seems to be in bed format but chr/start/end column names are missing
## VALIDATE REGIONS
## * Check if index is a string... PASS
## * Check index pattern... PASS
## * Check for missing values... PASS
## * Check for larger start position... PASS.
## * Check if zero based... PASS
## * Checking sort order... PASS
## * Checking for overlapping 'contiguous' regions... FAIL
## The input for object has overlapping features!
## This can cause unexpected results for some set operations.
## i.e. x <- bedr.merge.region(x)
## * Processing input (2): b
## CONVERT TO BED
## * Checking input type... PASS
## Input seems to be in bed format but chr/start/end column names are missing
## VALIDATE REGIONS
## * Check if index is a string... PASS
## * Check index pattern... PASS
## * Check for missing values... PASS
## * Check for larger start position... PASS.
## * Check if zero based... PASS
## * Checking sort order... FAIL
## The input for object is not *lexographically* ordered!
## This can cause unexpected results for some set operations.
## try: x <- bedr.sort.region(x)
## * Checking for overlapping 'contiguous' regions... FAIL
## The input for object has overlapping features!
## This can cause unexpected results for some set operations.
## i.e. x <- bedr.merge.region(x)
## bedtools closest -a /var/folders/rf/qrcw6ncj05z1pc_pq9xzw3540000gn/T//RtmpXK3I56/a_bc55c1f3227.bed -b /var/folders/rf/qrcw6ncj05z1pc_pq9xzw3540000gn/T//RtmpXK3I56/b_bc553f2d81a4.bed# Convert the gene name to ensg
gene_id <- read.table("../../../Reg_Evo_Primates/data/ENSG_GENE_HG19.csv", stringsAsFactors = FALSE, header=TRUE, sep = ",")
comb_kidney <- merge(closest_heart, gene_id, by.x = c("V8"), by.y = c("Gene"))
comb_kidney$ensg## [1] "ENSG00000081760" "ENSG00000198691" "ENSG00000100997"
## [4] "ENSG00000100997" "ENSG00000117148" "ENSG00000169717"
## [7] "ENSG00000169717" "ENSG00000139567" "ENSG00000145536"
## [10] "ENSG00000134917" "ENSG00000105963" "ENSG00000162104"
## [13] "ENSG00000164252" "ENSG00000063438" "ENSG00000063438"
## [16] "ENSG00000110711" "ENSG00000196581" "ENSG00000196581"
## [19] "ENSG00000179841" "ENSG00000053371" "ENSG00000151360"
## [22] "ENSG00000178038" "ENSG00000178038" "ENSG00000106927"
## [25] "ENSG00000159461" "ENSG00000089053" "ENSG00000138613"
## [28] "ENSG00000138613" "ENSG00000198576" "ENSG00000147799"
## [31] "ENSG00000116017" "ENSG00000029153" "ENSG00000029153"
## [34] "ENSG00000029153" "ENSG00000029153" "ENSG00000029153"
## [37] "ENSG00000117407" "ENSG00000117407" "ENSG00000117407"
## [40] "ENSG00000126775" "ENSG00000126775" "ENSG00000068650"
## [43] "ENSG00000068650" "ENSG00000068650" "ENSG00000068650"
## [46] "ENSG00000241837" "ENSG00000241837" "ENSG00000158321"
## [49] "ENSG00000158321" "ENSG00000183778" "ENSG00000183778"
## [52] "ENSG00000183778" "ENSG00000183778" "ENSG00000183778"
## [55] "ENSG00000128298" "ENSG00000125492" "ENSG00000125492"
## [58] "ENSG00000125124" "ENSG00000050820" "ENSG00000127152"
## [61] "ENSG00000127152" "ENSG00000110987" "ENSG00000110987"
## [64] "ENSG00000116128" "ENSG00000162373" "ENSG00000165626"
## [67] "ENSG00000165626" "ENSG00000122870" "ENSG00000122870"
## [70] "ENSG00000136717" "ENSG00000136717" "ENSG00000136717"
## [73] "ENSG00000136717" "ENSG00000136717" "ENSG00000136717"
## [76] "ENSG00000136717" "ENSG00000136717" "ENSG00000136717"
## [79] "ENSG00000136717" "ENSG00000136717" "ENSG00000136717"
## [82] "ENSG00000136717" "ENSG00000136717" "ENSG00000136717"
## [85] "ENSG00000136717" "ENSG00000136717" "ENSG00000136717"
## [88] "ENSG00000136717" "ENSG00000136717" "ENSG00000125845"
## [91] "ENSG00000101144" "ENSG00000104221" "ENSG00000165863"
## [94] "ENSG00000109944" "ENSG00000109944" "ENSG00000173064"
## [97] "ENSG00000235162" "ENSG00000089916" "ENSG00000089916"
## [100] "ENSG00000184601" "ENSG00000180336" "ENSG00000131943"
## [103] "ENSG00000175262" "ENSG00000128346" "ENSG00000185056"
## [106] "ENSG00000146521" "ENSG00000074410" "ENSG00000074410"
## [109] "ENSG00000158966" "ENSG00000198286" "ENSG00000130940"
## [112] "ENSG00000130940" "ENSG00000160200" "ENSG00000135736"
## [115] "ENSG00000168491" "ENSG00000168491" "ENSG00000109881"
## [118] "ENSG00000109881" "ENSG00000135127" "ENSG00000135127"
## [121] "ENSG00000135127" "ENSG00000110092" "ENSG00000138764"
## [124] "ENSG00000126353" "ENSG00000146731" "ENSG00000146731"
## [127] "ENSG00000112149" "ENSG00000112149" "ENSG00000198752"
## [130] "ENSG00000184661" "ENSG00000166589" "ENSG00000166589"
## [133] "ENSG00000166589" "ENSG00000166589" "ENSG00000179242"
## [136] "ENSG00000179242" "ENSG00000113100" "ENSG00000227767"
## [139] "ENSG00000153046" "ENSG00000101489" "ENSG00000101489"
## [142] "ENSG00000101489" "ENSG00000101489" "ENSG00000134873"
## [145] "ENSG00000134873" "ENSG00000159261" "ENSG00000169583"
## [148] "ENSG00000120885" "ENSG00000148842" "ENSG00000148842"
## [151] "ENSG00000148842" "ENSG00000149972" "ENSG00000149972"
## [154] "ENSG00000149972" "ENSG00000134871" "ENSG00000130635"
## [157] "ENSG00000165644" "ENSG00000198612" "ENSG00000198612"
## [160] "ENSG00000091704" "ENSG00000091704" "ENSG00000169372"
## [163] "ENSG00000146592" "ENSG00000146592" "ENSG00000160202"
## [166] "ENSG00000060069" "ENSG00000060069" "ENSG00000134030"
## [169] "ENSG00000134030" "ENSG00000153015" "ENSG00000153015"
## [172] "ENSG00000153015" "ENSG00000019186" "ENSG00000019186"
## [175] "ENSG00000132437" "ENSG00000079785" "ENSG00000141141"
## [178] "ENSG00000160049" "ENSG00000160049" "ENSG00000109016"
## [181] "ENSG00000100697" "ENSG00000151240" "ENSG00000150672"
## [184] "ENSG00000151208" "ENSG00000080845" "ENSG00000119689"
## [187] "ENSG00000119689" "ENSG00000119689" "ENSG00000119689"
## [190] "ENSG00000143006" "ENSG00000079805" "ENSG00000079805"
## [193] "ENSG00000079805" "ENSG00000079805" "ENSG00000079805"
## [196] "ENSG00000130226" "ENSG00000156162" "ENSG00000149636"
## [199] "ENSG00000149636" "ENSG00000149636" "ENSG00000047579"
## [202] "ENSG00000047579" "ENSG00000047579" "ENSG00000143507"
## [205] "ENSG00000143507" "ENSG00000229847" "ENSG00000229847"
## [208] "ENSG00000229847" "ENSG00000164778" "ENSG00000167280"
## [211] "ENSG00000104714" "ENSG00000119715" "ENSG00000119715"
## [214] "ENSG00000072840" "ENSG00000064655" "ENSG00000064655"
## [217] "ENSG00000088926" "ENSG00000168309" "ENSG00000168309"
## [220] "ENSG00000184731" "ENSG00000196814" "ENSG00000196814"
## [223] "ENSG00000154153" "ENSG00000189292" "ENSG00000182183"
## [226] "ENSG00000115363" "ENSG00000047662" "ENSG00000219438"
## [229] "ENSG00000168672" "ENSG00000083857" "ENSG00000083857"
## [232] "ENSG00000165140" "ENSG00000112787" "ENSG00000112787"
## [235] "ENSG00000085265" "ENSG00000126266" "ENSG00000066468"
## [238] "ENSG00000066468" "ENSG00000066468" "ENSG00000126500"
## [241] "ENSG00000075426" "ENSG00000164916" "ENSG00000114861"
## [244] "ENSG00000150893" "ENSG00000033170" "ENSG00000166206"
## [247] "ENSG00000116717" "ENSG00000116717" "ENSG00000116717"
## [250] "ENSG00000116717" "ENSG00000116717" "ENSG00000116717"
## [253] "ENSG00000143641" "ENSG00000182870" "ENSG00000130700"
## [256] "ENSG00000168505" "ENSG00000165702" "ENSG00000165702"
## [259] "ENSG00000167741" "ENSG00000167741" "ENSG00000074047"
## [262] "ENSG00000107249" "ENSG00000151948" "ENSG00000151948"
## [265] "ENSG00000137198" "ENSG00000156049" "ENSG00000197177"
## [268] "ENSG00000077585" "ENSG00000158292" "ENSG00000119714"
## [271] "ENSG00000178075" "ENSG00000125651" "ENSG00000077235"
## [274] "ENSG00000069812" "ENSG00000166135" "ENSG00000180448"
## [277] "ENSG00000108753" "ENSG00000108753" "ENSG00000108753"
## [280] "ENSG00000108753" "ENSG00000136720" "ENSG00000165868"
## [283] "ENSG00000165868" "ENSG00000003147" "ENSG00000003147"
## [286] "ENSG00000003147" "ENSG00000160223" "ENSG00000172201"
## [289] "ENSG00000172201" "ENSG00000134049" "ENSG00000117154"
## [292] "ENSG00000117154" "ENSG00000185950" "ENSG00000113430"
## [295] "ENSG00000113430" "ENSG00000105655" "ENSG00000105655"
## [298] "ENSG00000105655" "ENSG00000188385" "ENSG00000154118"
## [301] "ENSG00000107104" "ENSG00000102781" "ENSG00000189337"
## [304] "ENSG00000115041" "ENSG00000169427" "ENSG00000053918"
## [307] "ENSG00000134504" "ENSG00000167977" "ENSG00000127663"
## [310] "ENSG00000127663" "ENSG00000119537" "ENSG00000131149"
## [313] "ENSG00000136051" "ENSG00000257093" "ENSG00000122778"
## [316] "ENSG00000122778" "ENSG00000140950" "ENSG00000173214"
## [319] "ENSG00000130294" "ENSG00000066735" "ENSG00000066735"
## [322] "ENSG00000067082" "ENSG00000067082" "ENSG00000067082"
## [325] "ENSG00000067082" "ENSG00000150361" "ENSG00000197705"
## [328] "ENSG00000053747" "ENSG00000053747" "ENSG00000091136"
## [331] "ENSG00000168961" "ENSG00000168961" "ENSG00000168961"
## [334] "ENSG00000132130" "ENSG00000073350" "ENSG00000073350"
## [337] "ENSG00000073350" "ENSG00000103227" "ENSG00000103227"
## [340] "ENSG00000113368" "ENSG00000163380" "ENSG00000136944"
## [343] "ENSG00000136944" "ENSG00000136944" "ENSG00000167210"
## [346] "ENSG00000153395" "ENSG00000136141" "ENSG00000136141"
## [349] "ENSG00000136141" "ENSG00000146006" "ENSG00000146006"
## [352] "ENSG00000166159" "ENSG00000166159" "ENSG00000166159"
## [355] "ENSG00000166159" "ENSG00000166159" "ENSG00000166159"
## [358] "ENSG00000119681" "ENSG00000002822" "ENSG00000002822"
## [361] "ENSG00000002822" "ENSG00000178573" "ENSG00000178573"
## [364] "ENSG00000185022" "ENSG00000172469" "ENSG00000135525"
## [367] "ENSG00000135525" "ENSG00000135525" "ENSG00000138834"
## [370] "ENSG00000138834" "ENSG00000137337" "ENSG00000085871"
## [373] "ENSG00000085871" "ENSG00000085871" "ENSG00000085871"
## [376] "ENSG00000249567" "ENSG00000221323" "ENSG00000207588"
## [379] "ENSG00000105926" "ENSG00000158186" "ENSG00000182170"
## [382] "ENSG00000153944" "ENSG00000138823" "ENSG00000129422"
## [385] "ENSG00000129422" "ENSG00000132938" "ENSG00000133055"
## [388] "ENSG00000169994" "ENSG00000187556" "ENSG00000196498"
## [391] "ENSG00000196498" "ENSG00000196498" "ENSG00000196498"
## [394] "ENSG00000196498" "ENSG00000196498" "ENSG00000104419"
## [397] "ENSG00000104419" "ENSG00000104419" "ENSG00000104419"
## [400] "ENSG00000115286" "ENSG00000103154" "ENSG00000109320"
## [403] "ENSG00000109320" "ENSG00000148826" "ENSG00000148826"
## [406] "ENSG00000106410" "ENSG00000106410" "ENSG00000074771"
## [409] "ENSG00000170485" "ENSG00000214285" "ENSG00000105954"
## [412] "ENSG00000182667" "ENSG00000182667" "ENSG00000182667"
## [415] "ENSG00000115758" "ENSG00000205927" "ENSG00000183715"
## [418] "ENSG00000115947" "ENSG00000178602" "ENSG00000172818"
## [421] "ENSG00000125850" "ENSG00000099864" "ENSG00000099864"
## [424] "ENSG00000116183" "ENSG00000116183" "ENSG00000137819"
## [427] "ENSG00000102699" "ENSG00000242265" "ENSG00000242265"
## [430] "ENSG00000142655" "ENSG00000142655" "ENSG00000067057"
## [433] "ENSG00000144824" "ENSG00000165443" "ENSG00000105229"
## [436] "ENSG00000126822" "ENSG00000171680" "ENSG00000114554"
## [439] "ENSG00000114554" "ENSG00000114554" "ENSG00000141682"
## [442] "ENSG00000124225" "ENSG00000122512" "ENSG00000122512"
## [445] "ENSG00000168081" "ENSG00000014138" "ENSG00000166169"
## [448] "ENSG00000166169" "ENSG00000166169" "ENSG00000166169"
## [451] "ENSG00000106536" "ENSG00000106536" "ENSG00000162407"
## [454] "ENSG00000147535" "ENSG00000147535" "ENSG00000147535"
## [457] "ENSG00000154001" "ENSG00000170325" "ENSG00000170325"
## [460] "ENSG00000110851" "ENSG00000175785" "ENSG00000111725"
## [463] "ENSG00000106617" "ENSG00000065675" "ENSG00000065675"
## [466] "ENSG00000111218" "ENSG00000111218" "ENSG00000155066"
## [469] "ENSG00000155066" "ENSG00000155066" "ENSG00000205352"
## [472] "ENSG00000205352" "ENSG00000205352" "ENSG00000205352"
## [475] "ENSG00000167157" "ENSG00000163636" "ENSG00000169398"
## [478] "ENSG00000169398" "ENSG00000169398" "ENSG00000134644"
## [481] "ENSG00000134644" "ENSG00000112531" "ENSG00000112531"
## [484] "ENSG00000112531" "ENSG00000112531" "ENSG00000107560"
## [487] "ENSG00000168461" "ENSG00000141542" "ENSG00000017797"
## [490] "ENSG00000136828" "ENSG00000136828" "ENSG00000136828"
## [493] "ENSG00000185989" "ENSG00000165105" "ENSG00000122035"
## [496] "ENSG00000122965" "ENSG00000122965" "ENSG00000122965"
## [499] "ENSG00000160957" "ENSG00000143954" "ENSG00000143954"
## [502] "ENSG00000143954" "ENSG00000159788" "ENSG00000091844"
## [505] "ENSG00000158106" "ENSG00000183421" "ENSG00000139797"
## [508] "ENSG00000113269" "ENSG00000069667" "ENSG00000100784"
## [511] "ENSG00000100784" "ENSG00000141564" "ENSG00000141564"
## [514] "ENSG00000165526" "ENSG00000165526" "ENSG00000182010"
## [517] "ENSG00000198838" "ENSG00000198838" "ENSG00000256463"
## [520] "ENSG00000256463" "ENSG00000256463" "ENSG00000149021"
## [523] "ENSG00000136546" "ENSG00000136546" "ENSG00000146555"
## [526] "ENSG00000146555" "ENSG00000187764" "ENSG00000187764"
## [529] "ENSG00000163904" "ENSG00000152217" "ENSG00000183576"
## [532] "ENSG00000183576" "ENSG00000225383" "ENSG00000164690"
## [535] "ENSG00000142178" "ENSG00000159263" "ENSG00000159263"
## [538] "ENSG00000159263" "ENSG00000159263" "ENSG00000113504"
## [541] "ENSG00000152779" "ENSG00000162241" "ENSG00000162241"
## [544] "ENSG00000173262" "ENSG00000157765" "ENSG00000157765"
## [547] "ENSG00000160190" "ENSG00000143036" "ENSG00000143036"
## [550] "ENSG00000143036" "ENSG00000162426" "ENSG00000066230"
## [553] "ENSG00000173930" "ENSG00000166949" "ENSG00000127616"
## [556] "ENSG00000127616" "ENSG00000127616" "ENSG00000127616"
## [559] "ENSG00000188176" "ENSG00000048471" "ENSG00000172803"
## [562] "ENSG00000184557" "ENSG00000125398" "ENSG00000125398"
## [565] "ENSG00000185594" "ENSG00000187678" "ENSG00000187678"
## [568] "ENSG00000124783" "ENSG00000149418" "ENSG00000133121"
## [571] "ENSG00000133121" "ENSG00000102572" "ENSG00000152953"
## [574] "ENSG00000015592" "ENSG00000165730" "ENSG00000165730"
## [577] "ENSG00000165730" "ENSG00000165730" "ENSG00000198203"
## [580] "ENSG00000198203" "ENSG00000117614" "ENSG00000117614"
## [583] "ENSG00000131018" "ENSG00000131018" "ENSG00000078269"
## [586] "ENSG00000143028" "ENSG00000130699" "ENSG00000164691"
## [589] "ENSG00000164691" "ENSG00000164691" "ENSG00000167291"
## [592] "ENSG00000095383" "ENSG00000095383" "ENSG00000095383"
## [595] "ENSG00000095383" "ENSG00000095383" "ENSG00000095383"
## [598] "ENSG00000006638" "ENSG00000006638" "ENSG00000187621"
## [601] "ENSG00000160180" "ENSG00000163235" "ENSG00000163235"
## [604] "ENSG00000041988" "ENSG00000041988" "ENSG00000146426"
## [607] "ENSG00000106829" "ENSG00000155957" "ENSG00000144120"
## [610] "ENSG00000144120" "ENSG00000144120" "ENSG00000151353"
## [613] "ENSG00000151353" "ENSG00000214128" "ENSG00000160055"
## [616] "ENSG00000136205" "ENSG00000162341" "ENSG00000115705"
## [619] "ENSG00000115705" "ENSG00000126602" "ENSG00000167632"
## [622] "ENSG00000071575" "ENSG00000071575" "ENSG00000137699"
## [625] "ENSG00000100815" "ENSG00000103671" "ENSG00000119121"
## [628] "ENSG00000154743" "ENSG00000154743" "ENSG00000179981"
## [631] "ENSG00000182463" "ENSG00000121297" "ENSG00000214063"
## [634] "ENSG00000032389" "ENSG00000123607" "ENSG00000105948"
## [637] "ENSG00000105948" "ENSG00000105948" "ENSG00000100154"
## [640] "ENSG00000143367" "ENSG00000143367" "ENSG00000233608"
## [643] "ENSG00000184787" "ENSG00000184787" "ENSG00000184787"
## [646] "ENSG00000178473" "ENSG00000115446" "ENSG00000113763"
## [649] "ENSG00000101558" "ENSG00000101558" "ENSG00000129003"
## [652] "ENSG00000129003" "ENSG00000129003" "ENSG00000129003"
## [655] "ENSG00000176428" "ENSG00000185274" "ENSG00000185274"
## [658] "ENSG00000185274" "ENSG00000065268" "ENSG00000160193"
## [661] "ENSG00000103175" "ENSG00000198373" "ENSG00000076924"
## [664] "ENSG00000196584" "ENSG00000130733" "ENSG00000184828"
## [667] "ENSG00000213588" "ENSG00000014164" "ENSG00000175048"
## [670] "ENSG00000175048" "ENSG00000153786" "ENSG00000153786"
## [673] "ENSG00000156639" "ENSG00000156639" "ENSG00000133858"
## [676] "ENSG00000139800" "ENSG00000015171" "ENSG00000015171"
## [679] "ENSG00000102935" "ENSG00000102935" "ENSG00000183621"
## [682] "ENSG00000183621" "ENSG00000183621" "ENSG00000183621"
## [685] "ENSG00000183621" "ENSG00000183621" "ENSG00000183621"
## [688] "ENSG00000197857" "ENSG00000197857" "ENSG00000225614"
## [691] "ENSG00000225614" "ENSG00000171425" "ENSG00000157657"
## [694] "ENSG00000157657" "ENSG00000183779" "ENSG00000183779"# Convert the TSS to ensg as well
ref_gene_hg19_ensg <- merge(refGene_hg19_TSS, gene_id, by.x = c("V5"), by.y = c("Gene"))
all_ref_gene_hg19_ensg <- unique(ref_gene_hg19_ensg$ensg)
kidney_ref_gene <- all_ref_gene_hg19_ensg %in% comb_kidney$ensg
# Revisions- run GO
# Merge ENSG with true/false
test_gene <- as.numeric(as.vector(heart_ref_gene))
names(test_gene) <- all_ref_gene_hg19_ensg
# Run topGO
go_data <- new("topGOdata",
ontology = "BP",
allGenes = test_gene,
geneSel = function(allScore){
return(allScore > 0)
},
nodeSize = 5,
annotationFun = annFUN.org,
mapping = "org.Hs.eg.db",
ID = "ensembl")##
## Building most specific GOs .....## ( 11450 GO terms found. )##
## Build GO DAG topology ..........## ( 15456 GO terms and 36153 relations. )##
## Annotating nodes ...............## ( 14505 genes annotated to the GO terms. )# Perform enrichment test
go_test <- runTest(go_data, algorithm = "weight01", statistic = "fisher")##
## -- Weight01 Algorithm --
##
## the algorithm is scoring 1568 nontrivial nodes
## parameters:
## test statistic: fisher##
## Level 15: 1 nodes to be scored (0 eliminated genes)##
## Level 14: 11 nodes to be scored (0 eliminated genes)##
## Level 13: 29 nodes to be scored (103 eliminated genes)##
## Level 12: 50 nodes to be scored (298 eliminated genes)##
## Level 11: 72 nodes to be scored (2501 eliminated genes)##
## Level 10: 107 nodes to be scored (3937 eliminated genes)##
## Level 9: 161 nodes to be scored (5541 eliminated genes)##
## Level 8: 201 nodes to be scored (7251 eliminated genes)##
## Level 7: 245 nodes to be scored (8558 eliminated genes)##
## Level 6: 265 nodes to be scored (10615 eliminated genes)##
## Level 5: 217 nodes to be scored (12035 eliminated genes)##
## Level 4: 127 nodes to be scored (13480 eliminated genes)##
## Level 3: 63 nodes to be scored (13985 eliminated genes)##
## Level 2: 18 nodes to be scored (14177 eliminated genes)##
## Level 1: 1 nodes to be scored (14301 eliminated genes)go_table <- GenTable(go_data, weightFisher = go_test,
orderBy = "weightFisher", ranksOf = "weightFisher",
topNodes = sum(score(go_test) < .01))
go_table## GO.ID Term Annotated
## 1 GO:0009113 purine nucleobase biosynthetic process 12
## 2 GO:0033197 response to vitamin E 12
## 3 GO:0006351 transcription, DNA-templated 3277
## 4 GO:0032007 negative regulation of TOR signaling 40
## 5 GO:0043547 positive regulation of GTPase activity 354
## 6 GO:0060070 canonical Wnt signaling pathway 274
## 7 GO:1903078 positive regulation of protein localizat... 46
## 8 GO:0071320 cellular response to cAMP 47
## Significant Expected weightFisher
## 1 2 0.03 0.00046
## 2 2 0.03 0.00046
## 3 14 8.81 0.00408
## 4 2 0.11 0.00515
## 5 6 0.95 0.00540
## 6 4 0.74 0.00656
## 7 2 0.12 0.00677
## 8 2 0.13 0.00706goresults["GO:0098719"] ## $<NA>
## NULLLiver
# Find the heart hypo
heart_hypo <- human_chimp_heart_specific_25_conserved_H3K27_LIVER[which(human_chimp_heart_specific_25_conserved_H3K27_LIVER$V4 == "hypo" & human_chimp_heart_specific_25_conserved_H3K27_LIVER$V6 != "-1"),]
heart_hypo <- heart_hypo[complete.cases(heart_hypo), ]
# Find the closest gene
closest_heart <- bedr(input = list(a = heart_hypo[,1:3], b = refGene_hg19_TSS), method = "closest", check.chr = FALSE)## * Processing input (1): a
## CONVERT TO BED
## * Checking input type... PASS
## Input seems to be in bed format but chr/start/end column names are missing
## VALIDATE REGIONS
## * Check if index is a string... PASS
## * Check index pattern... PASS
## * Check for missing values... PASS
## * Check for larger start position... PASS.
## * Check if zero based... PASS
## * Checking sort order... PASS
## * Checking for overlapping 'contiguous' regions... FAIL
## The input for object has overlapping features!
## This can cause unexpected results for some set operations.
## i.e. x <- bedr.merge.region(x)
## * Processing input (2): b
## CONVERT TO BED
## * Checking input type... PASS
## Input seems to be in bed format but chr/start/end column names are missing
## VALIDATE REGIONS
## * Check if index is a string... PASS
## * Check index pattern... PASS
## * Check for missing values... PASS
## * Check for larger start position... PASS.
## * Check if zero based... PASS
## * Checking sort order... FAIL
## The input for object is not *lexographically* ordered!
## This can cause unexpected results for some set operations.
## try: x <- bedr.sort.region(x)
## * Checking for overlapping 'contiguous' regions... FAIL
## The input for object has overlapping features!
## This can cause unexpected results for some set operations.
## i.e. x <- bedr.merge.region(x)
## bedtools closest -a /var/folders/rf/qrcw6ncj05z1pc_pq9xzw3540000gn/T//RtmpXK3I56/a_bc556d78f000.bed -b /var/folders/rf/qrcw6ncj05z1pc_pq9xzw3540000gn/T//RtmpXK3I56/b_bc556f7b3abb.bed# Convert the gene name to ensg
gene_id <- read.table("../../../Reg_Evo_Primates/data/ENSG_GENE_HG19.csv", stringsAsFactors = FALSE, header=TRUE, sep = ",")
comb_kidney <- merge(closest_heart, gene_id, by.x = c("V8"), by.y = c("Gene"))
comb_kidney$ensg## [1] "ENSG00000121410" "ENSG00000114771" "ENSG00000114771"
## [4] "ENSG00000141338" "ENSG00000121270" "ENSG00000108846"
## [7] "ENSG00000108846" "ENSG00000108846" "ENSG00000108846"
## [10] "ENSG00000138075" "ENSG00000076555" "ENSG00000076555"
## [13] "ENSG00000181513" "ENSG00000181513" "ENSG00000181513"
## [16] "ENSG00000181513" "ENSG00000122729" "ENSG00000197142"
## [19] "ENSG00000183549" "ENSG00000143199" "ENSG00000143199"
## [22] "ENSG00000143199" "ENSG00000143199" "ENSG00000162104"
## [25] "ENSG00000183077" "ENSG00000183077" "ENSG00000183077"
## [28] "ENSG00000157985" "ENSG00000157985" "ENSG00000157985"
## [31] "ENSG00000165923" "ENSG00000165923" "ENSG00000165923"
## [34] "ENSG00000144891" "ENSG00000144891" "ENSG00000144891"
## [37] "ENSG00000144891" "ENSG00000172482" "ENSG00000113492"
## [40] "ENSG00000042286" "ENSG00000042286" "ENSG00000011243"
## [43] "ENSG00000106948" "ENSG00000198610" "ENSG00000198610"
## [46] "ENSG00000142208" "ENSG00000023330" "ENSG00000023330"
## [49] "ENSG00000170017" "ENSG00000170017" "ENSG00000170017"
## [52] "ENSG00000170017" "ENSG00000170017" "ENSG00000170017"
## [55] "ENSG00000170017" "ENSG00000170017" "ENSG00000033011"
## [58] "ENSG00000198796" "ENSG00000136383" "ENSG00000136383"
## [61] "ENSG00000162551" "ENSG00000162551" "ENSG00000162551"
## [64] "ENSG00000106927" "ENSG00000139344" "ENSG00000166126"
## [67] "ENSG00000166025" "ENSG00000166025" "ENSG00000116337"
## [70] "ENSG00000116337" "ENSG00000174945" "ENSG00000116194"
## [73] "ENSG00000116194" "ENSG00000198483" "ENSG00000154945"
## [76] "ENSG00000131620" "ENSG00000131620" "ENSG00000074855"
## [79] "ENSG00000042753" "ENSG00000042753" "ENSG00000132703"
## [82] "ENSG00000110243" "ENSG00000110243" "ENSG00000084674"
## [85] "ENSG00000175336" "ENSG00000118520" "ENSG00000118520"
## [88] "ENSG00000118520" "ENSG00000118520" "ENSG00000196843"
## [91] "ENSG00000133794" "ENSG00000133794" "ENSG00000133794"
## [94] "ENSG00000136950" "ENSG00000111339" "ENSG00000111339"
## [97] "ENSG00000100325" "ENSG00000100325" "ENSG00000141505"
## [100] "ENSG00000141505" "ENSG00000141505" "ENSG00000141505"
## [103] "ENSG00000161944" "ENSG00000161944" "ENSG00000161944"
## [106] "ENSG00000161944" "ENSG00000161944" "ENSG00000126522"
## [109] "ENSG00000126522" "ENSG00000126522" "ENSG00000169696"
## [112] "ENSG00000169696" "ENSG00000169696" "ENSG00000169136"
## [115] "ENSG00000169136" "ENSG00000157087" "ENSG00000157087"
## [118] "ENSG00000186009" "ENSG00000107518" "ENSG00000204842"
## [121] "ENSG00000158321" "ENSG00000119986" "ENSG00000109956"
## [124] "ENSG00000109956" "ENSG00000182272" "ENSG00000117411"
## [127] "ENSG00000156273" "ENSG00000175866" "ENSG00000175866"
## [130] "ENSG00000175866" "ENSG00000175866" "ENSG00000255056"
## [133] "ENSG00000172530" "ENSG00000172530" "ENSG00000172530"
## [136] "ENSG00000172530" "ENSG00000172530" "ENSG00000114200"
## [139] "ENSG00000153094" "ENSG00000153094" "ENSG00000153094"
## [142] "ENSG00000153094" "ENSG00000153094" "ENSG00000153094"
## [145] "ENSG00000153094" "ENSG00000153094" "ENSG00000153094"
## [148] "ENSG00000153094" "ENSG00000153094" "ENSG00000153094"
## [151] "ENSG00000153094" "ENSG00000153094" "ENSG00000153094"
## [154] "ENSG00000153094" "ENSG00000153094" "ENSG00000153094"
## [157] "ENSG00000121380" "ENSG00000121380" "ENSG00000100739"
## [160] "ENSG00000015475" "ENSG00000015475" "ENSG00000136717"
## [163] "ENSG00000136717" "ENSG00000136717" "ENSG00000136717"
## [166] "ENSG00000136717" "ENSG00000136717" "ENSG00000136717"
## [169] "ENSG00000136717" "ENSG00000136717" "ENSG00000136717"
## [172] "ENSG00000115760" "ENSG00000197299" "ENSG00000168487"
## [175] "ENSG00000168487" "ENSG00000168487" "ENSG00000168487"
## [178] "ENSG00000140299" "ENSG00000145919" "ENSG00000145919"
## [181] "ENSG00000176720" "ENSG00000234235" "ENSG00000214688"
## [184] "ENSG00000110696" "ENSG00000111412" "ENSG00000173064"
## [187] "ENSG00000214900" "ENSG00000128944" "ENSG00000128944"
## [190] "ENSG00000128944" "ENSG00000166780" "ENSG00000125149"
## [193] "ENSG00000125149" "ENSG00000174109" "ENSG00000185504"
## [196] "ENSG00000185504" "ENSG00000168675" "ENSG00000168675"
## [199] "ENSG00000168675" "ENSG00000168675" "ENSG00000168675"
## [202] "ENSG00000168675" "ENSG00000130813" "ENSG00000130813"
## [205] "ENSG00000130173" "ENSG00000168275" "ENSG00000159403"
## [208] "ENSG00000159403" "ENSG00000182326" "ENSG00000182326"
## [211] "ENSG00000166278" "ENSG00000166278" "ENSG00000166278"
## [214] "ENSG00000166278" "ENSG00000101474" "ENSG00000128254"
## [217] "ENSG00000215012" "ENSG00000100249" "ENSG00000115998"
## [220] "ENSG00000204128" "ENSG00000215217" "ENSG00000039537"
## [223] "ENSG00000146521" "ENSG00000244291" "ENSG00000170279"
## [226] "ENSG00000157131" "ENSG00000021852" "ENSG00000021852"
## [229] "ENSG00000021852" "ENSG00000176919" "ENSG00000113600"
## [232] "ENSG00000134508" "ENSG00000134508" "ENSG00000154040"
## [235] "ENSG00000154040" "ENSG00000154040" "ENSG00000154040"
## [238] "ENSG00000154040" "ENSG00000171735" "ENSG00000171735"
## [241] "ENSG00000171735" "ENSG00000171735" "ENSG00000042493"
## [244] "ENSG00000110888" "ENSG00000110888" "ENSG00000110888"
## [247] "ENSG00000110888" "ENSG00000110888" "ENSG00000213995"
## [250] "ENSG00000213995" "ENSG00000142273" "ENSG00000142273"
## [253] "ENSG00000139899" "ENSG00000139899" "ENSG00000160200"
## [256] "ENSG00000160200" "ENSG00000144648" "ENSG00000005059"
## [259] "ENSG00000160799" "ENSG00000144395" "ENSG00000165972"
## [262] "ENSG00000161573" "ENSG00000110092" "ENSG00000110092"
## [265] "ENSG00000177575" "ENSG00000177575" "ENSG00000177575"
## [268] "ENSG00000177575" "ENSG00000134061" "ENSG00000178562"
## [271] "ENSG00000178562" "ENSG00000178562" "ENSG00000110651"
## [274] "ENSG00000151465" "ENSG00000099804" "ENSG00000198752"
## [277] "ENSG00000149798" "ENSG00000170779" "ENSG00000170779"
## [280] "ENSG00000136807" "ENSG00000166446" "ENSG00000153879"
## [283] "ENSG00000153879" "ENSG00000241832" "ENSG00000149187"
## [286] "ENSG00000011523" "ENSG00000011523" "ENSG00000159398"
## [289] "ENSG00000159398" "ENSG00000087237" "ENSG00000243649"
## [292] "ENSG00000165410" "ENSG00000165410" "ENSG00000165410"
## [295] "ENSG00000016391" "ENSG00000016391" "ENSG00000085872"
## [298] "ENSG00000177830" "ENSG00000177830" "ENSG00000177830"
## [301] "ENSG00000177830" "ENSG00000177830" "ENSG00000110721"
## [304] "ENSG00000110721" "ENSG00000104879" "ENSG00000159261"
## [307] "ENSG00000159261" "ENSG00000125246" "ENSG00000091317"
## [310] "ENSG00000091317" "ENSG00000170293" "ENSG00000150656"
## [313] "ENSG00000133313" "ENSG00000168763" "ENSG00000168763"
## [316] "ENSG00000158158" "ENSG00000184144" "ENSG00000106078"
## [319] "ENSG00000196739" "ENSG00000118004" "ENSG00000118004"
## [322] "ENSG00000118004" "ENSG00000118004" "ENSG00000118004"
## [325] "ENSG00000118004" "ENSG00000131143" "ENSG00000178772"
## [328] "ENSG00000178772" "ENSG00000178772" "ENSG00000100884"
## [331] "ENSG00000157184" "ENSG00000167193" "ENSG00000167193"
## [334] "ENSG00000178585" "ENSG00000178585" "ENSG00000040531"
## [337] "ENSG00000040531" "ENSG00000111249" "ENSG00000111249"
## [340] "ENSG00000154639" "ENSG00000154639" "ENSG00000154639"
## [343] "ENSG00000154639" "ENSG00000154639" "ENSG00000166347"
## [346] "ENSG00000166347" "ENSG00000166347" "ENSG00000166347"
## [349] "ENSG00000166347" "ENSG00000166347" "ENSG00000166394"
## [352] "ENSG00000140505" "ENSG00000135929" "ENSG00000135929"
## [355] "ENSG00000135929" "ENSG00000138115" "ENSG00000138115"
## [358] "ENSG00000138115" "ENSG00000138115" "ENSG00000106258"
## [361] "ENSG00000106258" "ENSG00000106258" "ENSG00000106258"
## [364] "ENSG00000106258" "ENSG00000106258" "ENSG00000106258"
## [367] "ENSG00000106258" "ENSG00000106258" "ENSG00000106258"
## [370] "ENSG00000106258" "ENSG00000106258" "ENSG00000106258"
## [373] "ENSG00000106258" "ENSG00000106258" "ENSG00000106258"
## [376] "ENSG00000186115" "ENSG00000171954" "ENSG00000171954"
## [379] "ENSG00000186529" "ENSG00000186529" "ENSG00000186529"
## [382] "ENSG00000164488" "ENSG00000112977" "ENSG00000172992"
## [385] "ENSG00000043093" "ENSG00000132437" "ENSG00000100201"
## [388] "ENSG00000100201" "ENSG00000141141" "ENSG00000100150"
## [391] "ENSG00000100150" "ENSG00000077044" "ENSG00000102967"
## [394] "ENSG00000181192" "ENSG00000181192" "ENSG00000067596"
## [397] "ENSG00000184047" "ENSG00000184047" "ENSG00000184047"
## [400] "ENSG00000184047" "ENSG00000258498" "ENSG00000258498"
## [403] "ENSG00000258498" "ENSG00000066084" "ENSG00000108176"
## [406] "ENSG00000108176" "ENSG00000163687" "ENSG00000163687"
## [409] "ENSG00000167130" "ENSG00000167130" "ENSG00000167130"
## [412] "ENSG00000184845" "ENSG00000143507" "ENSG00000161326"
## [415] "ENSG00000108861" "ENSG00000107404" "ENSG00000105204"
## [418] "ENSG00000105204" "ENSG00000105204" "ENSG00000123179"
## [421] "ENSG00000134463" "ENSG00000229715" "ENSG00000229715"
## [424] "ENSG00000142634" "ENSG00000169242" "ENSG00000169242"
## [427] "ENSG00000099617" "ENSG00000172889" "ENSG00000172889"
## [430] "ENSG00000146648" "ENSG00000146648" "ENSG00000146648"
## [433] "ENSG00000146648" "ENSG00000171570" "ENSG00000173812"
## [436] "ENSG00000106263" "ENSG00000106263" "ENSG00000066044"
## [439] "ENSG00000196361" "ENSG00000196361" "ENSG00000155849"
## [442] "ENSG00000155849" "ENSG00000155849" "ENSG00000134759"
## [445] "ENSG00000134759" "ENSG00000134759" "ENSG00000134759"
## [448] "ENSG00000134759" "ENSG00000134759" "ENSG00000134759"
## [451] "ENSG00000160963" "ENSG00000167136" "ENSG00000116016"
## [454] "ENSG00000182580" "ENSG00000196411" "ENSG00000196411"
## [457] "ENSG00000130427" "ENSG00000113719" "ENSG00000086619"
## [460] "ENSG00000139684" "ENSG00000196405" "ENSG00000205436"
## [463] "ENSG00000107371" "ENSG00000107371" "ENSG00000182197"
## [466] "ENSG00000092820" "ENSG00000126218" "ENSG00000180210"
## [469] "ENSG00000159784" "ENSG00000109794" "ENSG00000152102"
## [472] "ENSG00000185442" "ENSG00000188916" "ENSG00000189319"
## [475] "ENSG00000133477" "ENSG00000188522" "ENSG00000176853"
## [478] "ENSG00000169710" "ENSG00000177294" "ENSG00000100225"
## [481] "ENSG00000100225" "ENSG00000171557" "ENSG00000171557"
## [484] "ENSG00000115226" "ENSG00000160097" "ENSG00000160097"
## [487] "ENSG00000170802" "ENSG00000106701" "ENSG00000106701"
## [490] "ENSG00000070404" "ENSG00000160282" "ENSG00000160282"
## [493] "ENSG00000107164" "ENSG00000165060" "ENSG00000165060"
## [496] "ENSG00000165060" "ENSG00000163251" "ENSG00000131482"
## [499] "ENSG00000131482" "ENSG00000154252" "ENSG00000108479"
## [502] "ENSG00000106648" "ENSG00000106648" "ENSG00000128310"
## [505] "ENSG00000130005" "ENSG00000130005" "ENSG00000111640"
## [508] "ENSG00000213512" "ENSG00000213512" "ENSG00000106633"
## [511] "ENSG00000106633" "ENSG00000084734" "ENSG00000023909"
## [514] "ENSG00000178795" "ENSG00000141098" "ENSG00000141098"
## [517] "ENSG00000141098" "ENSG00000151892" "ENSG00000115486"
## [520] "ENSG00000115486" "ENSG00000157017" "ENSG00000157017"
## [523] "ENSG00000157017" "ENSG00000157017" "ENSG00000157017"
## [526] "ENSG00000157017" "ENSG00000157017" "ENSG00000157017"
## [529] "ENSG00000157017" "ENSG00000157017" "ENSG00000137960"
## [532] "ENSG00000151948" "ENSG00000016864" "ENSG00000016864"
## [535] "ENSG00000203972" "ENSG00000196743" "ENSG00000196743"
## [538] "ENSG00000112699" "ENSG00000114349" "ENSG00000114349"
## [541] "ENSG00000100522" "ENSG00000185245" "ENSG00000119927"
## [544] "ENSG00000119927" "ENSG00000119927" "ENSG00000119927"
## [547] "ENSG00000112293" "ENSG00000132975" "ENSG00000180758"
## [550] "ENSG00000166123" "ENSG00000176153" "ENSG00000176153"
## [553] "ENSG00000176153" "ENSG00000075240" "ENSG00000083307"
## [556] "ENSG00000139835" "ENSG00000100577" "ENSG00000077809"
## [559] "ENSG00000077809" "ENSG00000077809" "ENSG00000077809"
## [562] "ENSG00000077809" "ENSG00000101181" "ENSG00000169919"
## [565] "ENSG00000130600" "ENSG00000148702" "ENSG00000148702"
## [568] "ENSG00000148702" "ENSG00000130956" "ENSG00000084110"
## [571] "ENSG00000084110" "ENSG00000084110" "ENSG00000115677"
## [574] "ENSG00000115677" "ENSG00000122557" "ENSG00000114315"
## [577] "ENSG00000114315" "ENSG00000168509" "ENSG00000168509"
## [580] "ENSG00000168509" "ENSG00000168509" "ENSG00000109758"
## [583] "ENSG00000137133" "ENSG00000100084" "ENSG00000108924"
## [586] "ENSG00000117305" "ENSG00000117305" "ENSG00000115756"
## [589] "ENSG00000158104" "ENSG00000110169" "ENSG00000110169"
## [592] "ENSG00000113905" "ENSG00000113905" "ENSG00000005700"
## [595] "ENSG00000115738" "ENSG00000142166" "ENSG00000142166"
## [598] "ENSG00000115457" "ENSG00000243646" "ENSG00000115590"
## [601] "ENSG00000115590" "ENSG00000196083" "ENSG00000196083"
## [604] "ENSG00000196083" "ENSG00000196083" "ENSG00000196083"
## [607] "ENSG00000196083" "ENSG00000196083" "ENSG00000196083"
## [610] "ENSG00000196083" "ENSG00000196083" "ENSG00000196083"
## [613] "ENSG00000196083" "ENSG00000196083" "ENSG00000196083"
## [616] "ENSG00000196083" "ENSG00000196083" "ENSG00000196083"
## [619] "ENSG00000196083" "ENSG00000196083" "ENSG00000196083"
## [622] "ENSG00000196083" "ENSG00000196083" "ENSG00000196083"
## [625] "ENSG00000196083" "ENSG00000136689" "ENSG00000103522"
## [628] "ENSG00000103522" "ENSG00000100385" "ENSG00000203485"
## [631] "ENSG00000203485" "ENSG00000203485" "ENSG00000122641"
## [634] "ENSG00000148384" "ENSG00000254647" "ENSG00000254647"
## [637] "ENSG00000254647" "ENSG00000129965" "ENSG00000129965"
## [640] "ENSG00000186480" "ENSG00000186480" "ENSG00000186480"
## [643] "ENSG00000186480" "ENSG00000186480" "ENSG00000186480"
## [646] "ENSG00000125629" "ENSG00000134070" "ENSG00000134070"
## [649] "ENSG00000172183" "ENSG00000078747" "ENSG00000078747"
## [652] "ENSG00000078747" "ENSG00000083457" "ENSG00000083457"
## [655] "ENSG00000162267" "ENSG00000055955" "ENSG00000055955"
## [658] "ENSG00000100605" "ENSG00000100605" "ENSG00000100605"
## [661] "ENSG00000205726" "ENSG00000205726" "ENSG00000163166"
## [664] "ENSG00000009765" "ENSG00000009765" "ENSG00000009765"
## [667] "ENSG00000140044" "ENSG00000154118" "ENSG00000102781"
## [670] "ENSG00000176407" "ENSG00000069424" "ENSG00000158445"
## [673] "ENSG00000124780" "ENSG00000169427" "ENSG00000188997"
## [676] "ENSG00000089094" "ENSG00000089094" "ENSG00000107077"
## [679] "ENSG00000107077" "ENSG00000166783" "ENSG00000166783"
## [682] "ENSG00000166783" "ENSG00000176542" "ENSG00000125337"
## [685] "ENSG00000125337" "ENSG00000118922" "ENSG00000129911"
## [688] "ENSG00000128607" "ENSG00000128607" "ENSG00000025800"
## [691] "ENSG00000185896" "ENSG00000002549" "ENSG00000107929"
## [694] "ENSG00000086730" "ENSG00000086730" "ENSG00000086730"
## [697] "ENSG00000213398" "ENSG00000164406" "ENSG00000164406"
## [700] "ENSG00000145826" "ENSG00000116977" "ENSG00000072163"
## [703] "ENSG00000166035" "ENSG00000166035" "ENSG00000166035"
## [706] "ENSG00000101670" "ENSG00000189067" "ENSG00000189067"
## [709] "ENSG00000162761" "ENSG00000102910" "ENSG00000134324"
## [712] "ENSG00000134324" "ENSG00000070018" "ENSG00000175489"
## [715] "ENSG00000160233" "ENSG00000124831" "ENSG00000124831"
## [718] "ENSG00000124831" "ENSG00000124831" "ENSG00000143669"
## [721] "ENSG00000197063" "ENSG00000145050" "ENSG00000101460"
## [724] "ENSG00000076984" "ENSG00000119487" "ENSG00000119487"
## [727] "ENSG00000119487" "ENSG00000119487" "ENSG00000119487"
## [730] "ENSG00000119487" "ENSG00000099785" "ENSG00000099785"
## [733] "ENSG00000099785" "ENSG00000009724" "ENSG00000009724"
## [736] "ENSG00000197971" "ENSG00000197971" "ENSG00000197971"
## [739] "ENSG00000197971" "ENSG00000126217" "ENSG00000180398"
## [742] "ENSG00000180398" "ENSG00000128285" "ENSG00000090674"
## [745] "ENSG00000135679" "ENSG00000135679" "ENSG00000135679"
## [748] "ENSG00000135679" "ENSG00000108510" "ENSG00000152127"
## [751] "ENSG00000170430" "ENSG00000199065" "ENSG00000199065"
## [754] "ENSG00000199065" "ENSG00000221039" "ENSG00000221063"
## [757] "ENSG00000208017" "ENSG00000199075" "ENSG00000207727"
## [760] "ENSG00000207875" "ENSG00000207875" "ENSG00000130382"
## [763] "ENSG00000115648" "ENSG00000115648" "ENSG00000009950"
## [766] "ENSG00000009950" "ENSG00000009950" "ENSG00000009950"
## [769] "ENSG00000108960" "ENSG00000173269" "ENSG00000075643"
## [772] "ENSG00000166391" "ENSG00000166391" "ENSG00000107186"
## [775] "ENSG00000107186" "ENSG00000107186" "ENSG00000214026"
## [778] "ENSG00000173867" "ENSG00000173531" "ENSG00000149480"
## [781] "ENSG00000122085" "ENSG00000122085" "ENSG00000122085"
## [784] "ENSG00000122085" "ENSG00000132613" "ENSG00000110921"
## [787] "ENSG00000110921" "ENSG00000172927" "ENSG00000172927"
## [790] "ENSG00000106436" "ENSG00000065534" "ENSG00000065534"
## [793] "ENSG00000065534" "ENSG00000065534" "ENSG00000091536"
## [796] "ENSG00000091536" "ENSG00000236242" "ENSG00000139597"
## [799] "ENSG00000139597" "ENSG00000008130" "ENSG00000008130"
## [802] "ENSG00000152620" "ENSG00000141562" "ENSG00000141562"
## [805] "ENSG00000141562" "ENSG00000166833" "ENSG00000255043"
## [808] "ENSG00000255043" "ENSG00000255043" "ENSG00000196498"
## [811] "ENSG00000196498" "ENSG00000196498" "ENSG00000166579"
## [814] "ENSG00000166579" "ENSG00000151366" "ENSG00000151366"
## [817] "ENSG00000151366" "ENSG00000165802" "ENSG00000165802"
## [820] "ENSG00000165802" "ENSG00000165802" "ENSG00000165802"
## [823] "ENSG00000173848" "ENSG00000107954" "ENSG00000100906"
## [826] "ENSG00000104825" "ENSG00000104825" "ENSG00000101004"
## [829] "ENSG00000163293" "ENSG00000172548" "ENSG00000172548"
## [832] "ENSG00000136783" "ENSG00000169251" "ENSG00000169251"
## [835] "ENSG00000166741" "ENSG00000015520" "ENSG00000015520"
## [838] "ENSG00000015520" "ENSG00000015520" "ENSG00000148734"
## [841] "ENSG00000242349" "ENSG00000131910" "ENSG00000012504"
## [844] "ENSG00000012504" "ENSG00000012504" "ENSG00000012504"
## [847] "ENSG00000012504" "ENSG00000012504" "ENSG00000012504"
## [850] "ENSG00000012504" "ENSG00000144852" "ENSG00000144852"
## [853] "ENSG00000144852" "ENSG00000143257" "ENSG00000143257"
## [856] "ENSG00000143257" "ENSG00000143257" "ENSG00000143257"
## [859] "ENSG00000143257" "ENSG00000143257" "ENSG00000143257"
## [862] "ENSG00000143257" "ENSG00000143257" "ENSG00000143257"
## [865] "ENSG00000143257" "ENSG00000143257" "ENSG00000143257"
## [868] "ENSG00000143257" "ENSG00000143257" "ENSG00000143257"
## [871] "ENSG00000143257" "ENSG00000143257" "ENSG00000143257"
## [874] "ENSG00000143257" "ENSG00000143257" "ENSG00000143257"
## [877] "ENSG00000143257" "ENSG00000143257" "ENSG00000143257"
## [880] "ENSG00000143257" "ENSG00000143257" "ENSG00000143257"
## [883] "ENSG00000143257" "ENSG00000113580" "ENSG00000113580"
## [886] "ENSG00000113580" "ENSG00000113580" "ENSG00000113580"
## [889] "ENSG00000113580" "ENSG00000113580" "ENSG00000113580"
## [892] "ENSG00000113580" "ENSG00000113580" "ENSG00000113580"
## [895] "ENSG00000113580" "ENSG00000113580" "ENSG00000113580"
## [898] "ENSG00000113580" "ENSG00000113580" "ENSG00000113580"
## [901] "ENSG00000113580" "ENSG00000113580" "ENSG00000113580"
## [904] "ENSG00000113580" "ENSG00000113580" "ENSG00000116833"
## [907] "ENSG00000116833" "ENSG00000116833" "ENSG00000116833"
## [910] "ENSG00000116833" "ENSG00000116833" "ENSG00000180530"
## [913] "ENSG00000171119" "ENSG00000151413" "ENSG00000090273"
## [916] "ENSG00000168101" "ENSG00000168101" "ENSG00000168101"
## [919] "ENSG00000168101" "ENSG00000132182" "ENSG00000125450"
## [922] "ENSG00000176046" "ENSG00000176046" "ENSG00000065154"
## [925] "ENSG00000065154" "ENSG00000180304" "ENSG00000111325"
## [928] "ENSG00000060491" "ENSG00000119547" "ENSG00000119547"
## [931] "ENSG00000224855" "ENSG00000175619" "ENSG00000140961"
## [934] "ENSG00000185624" "ENSG00000179364" "ENSG00000100266"
## [937] "ENSG00000006712" "ENSG00000006712" "ENSG00000006712"
## [940] "ENSG00000140694" "ENSG00000140694" "ENSG00000140694"
## [943] "ENSG00000140694" "ENSG00000140694" "ENSG00000140694"
## [946] "ENSG00000140694" "ENSG00000140694" "ENSG00000140694"
## [949] "ENSG00000111224" "ENSG00000185630" "ENSG00000185630"
## [952] "ENSG00000185630" "ENSG00000167081" "ENSG00000173599"
## [955] "ENSG00000166228" "ENSG00000175198" "ENSG00000175198"
## [958] "ENSG00000175198" "ENSG00000114054" "ENSG00000114054"
## [961] "ENSG00000140479" "ENSG00000140479" "ENSG00000140479"
## [964] "ENSG00000140479" "ENSG00000140479" "ENSG00000140479"
## [967] "ENSG00000140479" "ENSG00000140479" "ENSG00000169174"
## [970] "ENSG00000116005" "ENSG00000106244" "ENSG00000113721"
## [973] "ENSG00000113721" "ENSG00000155660" "ENSG00000107438"
## [976] "ENSG00000133027" "ENSG00000133027" "ENSG00000124299"
## [979] "ENSG00000124299" "ENSG00000124299" "ENSG00000124299"
## [982] "ENSG00000124299" "ENSG00000124299" "ENSG00000161031"
## [985] "ENSG00000107537" "ENSG00000060642" "ENSG00000155629"
## [988] "ENSG00000155629" "ENSG00000174238" "ENSG00000090975"
## [991] "ENSG00000178385" "ENSG00000076356" "ENSG00000115946"
## [994] "ENSG00000101346" "ENSG00000101346" "ENSG00000127948"
## [997] "ENSG00000168938" "ENSG00000168938" "ENSG00000108179"
## [1000] "ENSG00000118898" "ENSG00000219607" "ENSG00000137713"
## [1003] "ENSG00000137713" "ENSG00000137713" "ENSG00000137713"
## [1006] "ENSG00000137713" "ENSG00000066027" "ENSG00000066027"
## [1009] "ENSG00000066027" "ENSG00000100239" "ENSG00000100239"
## [1012] "ENSG00000100239" "ENSG00000100239" "ENSG00000110075"
## [1015] "ENSG00000110075" "ENSG00000110075" "ENSG00000110075"
## [1018] "ENSG00000110075" "ENSG00000110075" "ENSG00000147596"
## [1021] "ENSG00000116690" "ENSG00000116690" "ENSG00000116690"
## [1024] "ENSG00000116690" "ENSG00000131791" "ENSG00000154229"
## [1027] "ENSG00000067606" "ENSG00000101000" "ENSG00000117707"
## [1030] "ENSG00000117707" "ENSG00000126231" "ENSG00000126231"
## [1033] "ENSG00000161542" "ENSG00000161542" "ENSG00000248405"
## [1036] "ENSG00000080815" "ENSG00000080815" "ENSG00000159352"
## [1039] "ENSG00000180822" "ENSG00000180822" "ENSG00000180822"
## [1042] "ENSG00000124212" "ENSG00000206527" "ENSG00000070159"
## [1045] "ENSG00000070159" "ENSG00000070159" "ENSG00000070159"
## [1048] "ENSG00000060656" "ENSG00000060656" "ENSG00000060656"
## [1051] "ENSG00000060656" "ENSG00000060656" "ENSG00000060656"
## [1054] "ENSG00000060656" "ENSG00000060656" "ENSG00000136045"
## [1057] "ENSG00000136045" "ENSG00000165661" "ENSG00000213339"
## [1060] "ENSG00000172007" "ENSG00000172007" "ENSG00000172007"
## [1063] "ENSG00000160271" "ENSG00000108961" "ENSG00000108961"
## [1066] "ENSG00000108961" "ENSG00000173166" "ENSG00000173166"
## [1069] "ENSG00000105122" "ENSG00000108551" "ENSG00000108551"
## [1072] "ENSG00000165105" "ENSG00000100320" "ENSG00000100320"
## [1075] "ENSG00000184863" "ENSG00000138207" "ENSG00000139547"
## [1078] "ENSG00000139547" "ENSG00000174236" "ENSG00000173156"
## [1081] "ENSG00000111785" "ENSG00000167705" "ENSG00000108830"
## [1084] "ENSG00000108830" "ENSG00000178828" "ENSG00000132383"
## [1087] "ENSG00000125844" "ENSG00000125844" "ENSG00000160214"
## [1090] "ENSG00000163825" "ENSG00000163825" "ENSG00000163602"
## [1093] "ENSG00000020577" "ENSG00000020577" "ENSG00000101347"
## [1096] "ENSG00000123453" "ENSG00000168077" "ENSG00000168077"
## [1099] "ENSG00000073060" "ENSG00000073060" "ENSG00000073060"
## [1102] "ENSG00000073060" "ENSG00000073060" "ENSG00000073060"
## [1105] "ENSG00000143653" "ENSG00000100012" "ENSG00000100012"
## [1108] "ENSG00000100012" "ENSG00000100012" "ENSG00000103184"
## [1111] "ENSG00000254154" "ENSG00000254154" "ENSG00000065665"
## [1114] "ENSG00000065665" "ENSG00000065665" "ENSG00000065665"
## [1117] "ENSG00000065665" "ENSG00000025796" "ENSG00000187742"
## [1120] "ENSG00000187742" "ENSG00000179918" "ENSG00000184640"
## [1123] "ENSG00000197249" "ENSG00000196136" "ENSG00000100665"
## [1126] "ENSG00000117601" "ENSG00000117601" "ENSG00000183576"
## [1129] "ENSG00000183576" "ENSG00000144040" "ENSG00000144040"
## [1132] "ENSG00000144040" "ENSG00000104205" "ENSG00000104205"
## [1135] "ENSG00000130147" "ENSG00000107957" "ENSG00000105251"
## [1138] "ENSG00000105251" "ENSG00000138606" "ENSG00000164690"
## [1141] "ENSG00000164690" "ENSG00000138771" "ENSG00000112246"
## [1144] "ENSG00000169375" "ENSG00000096717" "ENSG00000124523"
## [1147] "ENSG00000141526" "ENSG00000170190" "ENSG00000112337"
## [1150] "ENSG00000112337" "ENSG00000112337" "ENSG00000146039"
## [1153] "ENSG00000110436" "ENSG00000168575" "ENSG00000175003"
## [1156] "ENSG00000175003" "ENSG00000175003" "ENSG00000175003"
## [1159] "ENSG00000149742" "ENSG00000148339" "ENSG00000171612"
## [1162] "ENSG00000171612" "ENSG00000171612" "ENSG00000181035"
## [1165] "ENSG00000140107" "ENSG00000140284" "ENSG00000140284"
## [1168] "ENSG00000083807" "ENSG00000197496" "ENSG00000173262"
## [1171] "ENSG00000196660" "ENSG00000196660" "ENSG00000196660"
## [1174] "ENSG00000196660" "ENSG00000130958" "ENSG00000127526"
## [1177] "ENSG00000138449" "ENSG00000162426" "ENSG00000022567"
## [1180] "ENSG00000022567" "ENSG00000076351" "ENSG00000076351"
## [1183] "ENSG00000139508" "ENSG00000139508" "ENSG00000111181"
## [1186] "ENSG00000111181" "ENSG00000111181" "ENSG00000130876"
## [1189] "ENSG00000130876" "ENSG00000103257" "ENSG00000137834"
## [1192] "ENSG00000137834" "ENSG00000137834" "ENSG00000137834"
## [1195] "ENSG00000103056" "ENSG00000103056" "ENSG00000198742"
## [1198] "ENSG00000198742" "ENSG00000198742" "ENSG00000165684"
## [1201] "ENSG00000201902" "ENSG00000104852" "ENSG00000147481"
## [1204] "ENSG00000142168" "ENSG00000142168" "ENSG00000184985"
## [1207] "ENSG00000184985" "ENSG00000137642" "ENSG00000100146"
## [1210] "ENSG00000153498" "ENSG00000227213" "ENSG00000006282"
## [1213] "ENSG00000006282" "ENSG00000006282" "ENSG00000186583"
## [1216] "ENSG00000186583" "ENSG00000204710" "ENSG00000100014"
## [1219] "ENSG00000072080" "ENSG00000136158" "ENSG00000162032"
## [1222] "ENSG00000173898" "ENSG00000137877" "ENSG00000049319"
## [1225] "ENSG00000182934" "ENSG00000182934" "ENSG00000147488"
## [1228] "ENSG00000166444" "ENSG00000073849" "ENSG00000070731"
## [1231] "ENSG00000133121" "ENSG00000166888" "ENSG00000115107"
## [1234] "ENSG00000115107" "ENSG00000115107" "ENSG00000115107"
## [1237] "ENSG00000115107" "ENSG00000115107" "ENSG00000118046"
## [1240] "ENSG00000204344" "ENSG00000116030" "ENSG00000116030"
## [1243] "ENSG00000116030" "ENSG00000116030" "ENSG00000116030"
## [1246] "ENSG00000116030" "ENSG00000100242" "ENSG00000196235"
## [1249] "ENSG00000173227" "ENSG00000177156" "ENSG00000143374"
## [1252] "ENSG00000135111" "ENSG00000135111" "ENSG00000135111"
## [1255] "ENSG00000135111" "ENSG00000135111" "ENSG00000135111"
## [1258] "ENSG00000011007" "ENSG00000163792" "ENSG00000163792"
## [1261] "ENSG00000148737" "ENSG00000148737" "ENSG00000148737"
## [1264] "ENSG00000148737" "ENSG00000148737" "ENSG00000148737"
## [1267] "ENSG00000148737" "ENSG00000148737" "ENSG00000148737"
## [1270] "ENSG00000148737" "ENSG00000148737" "ENSG00000148737"
## [1273] "ENSG00000148737" "ENSG00000148737" "ENSG00000148737"
## [1276] "ENSG00000148737" "ENSG00000148737" "ENSG00000148737"
## [1279] "ENSG00000148737" "ENSG00000148737" "ENSG00000148737"
## [1282] "ENSG00000148737" "ENSG00000148737" "ENSG00000148737"
## [1285] "ENSG00000148737" "ENSG00000148737" "ENSG00000148737"
## [1288] "ENSG00000148737" "ENSG00000148737" "ENSG00000148737"
## [1291] "ENSG00000148737" "ENSG00000148737" "ENSG00000148737"
## [1294] "ENSG00000148737" "ENSG00000148737" "ENSG00000148737"
## [1297] "ENSG00000148737" "ENSG00000148737" "ENSG00000148737"
## [1300] "ENSG00000148737" "ENSG00000148737" "ENSG00000148737"
## [1303] "ENSG00000148737" "ENSG00000148737" "ENSG00000148737"
## [1306] "ENSG00000148737" "ENSG00000148737" "ENSG00000148737"
## [1309] "ENSG00000148737" "ENSG00000148737" "ENSG00000148737"
## [1312] "ENSG00000148737" "ENSG00000101190" "ENSG00000100109"
## [1315] "ENSG00000100109" "ENSG00000106327" "ENSG00000072274"
## [1318] "ENSG00000125780" "ENSG00000125780" "ENSG00000101158"
## [1321] "ENSG00000079134" "ENSG00000172009" "ENSG00000090534"
## [1324] "ENSG00000090534" "ENSG00000090534" "ENSG00000187720"
## [1327] "ENSG00000221995" "ENSG00000104980" "ENSG00000100234"
## [1330] "ENSG00000150455" "ENSG00000150455" "ENSG00000196781"
## [1333] "ENSG00000140332" "ENSG00000140332" "ENSG00000140332"
## [1336] "ENSG00000135926" "ENSG00000134291" "ENSG00000134291"
## [1339] "ENSG00000134291" "ENSG00000134291" "ENSG00000168936"
## [1342] "ENSG00000168936" "ENSG00000064545" "ENSG00000064545"
## [1345] "ENSG00000106565" "ENSG00000106565" "ENSG00000106565"
## [1348] "ENSG00000187824" "ENSG00000204278" "ENSG00000204278"
## [1351] "ENSG00000204278" "ENSG00000149582" "ENSG00000149582"
## [1354] "ENSG00000149582" "ENSG00000149582" "ENSG00000149582"
## [1357] "ENSG00000149582" "ENSG00000109133" "ENSG00000109133"
## [1360] "ENSG00000177042" "ENSG00000177042" "ENSG00000109084"
## [1363] "ENSG00000187045" "ENSG00000109079" "ENSG00000185361"
## [1366] "ENSG00000028137" "ENSG00000141232" "ENSG00000141232"
## [1369] "ENSG00000136816" "ENSG00000188001" "ENSG00000056558"
## [1372] "ENSG00000131653" "ENSG00000131653" "ENSG00000173334"
## [1375] "ENSG00000173334" "ENSG00000173334" "ENSG00000101255"
## [1378] "ENSG00000234127" "ENSG00000234127" "ENSG00000169871"
## [1381] "ENSG00000144481" "ENSG00000144481" "ENSG00000011105"
## [1384] "ENSG00000011105" "ENSG00000235217" "ENSG00000172425"
## [1387] "ENSG00000100304" "ENSG00000120440" "ENSG00000124120"
## [1390] "ENSG00000124120" "ENSG00000124120" "ENSG00000118271"
## [1393] "ENSG00000136295" "ENSG00000198431" "ENSG00000143569"
## [1396] "ENSG00000078140" "ENSG00000078140" "ENSG00000078140"
## [1399] "ENSG00000169139" "ENSG00000168246" "ENSG00000162543"
## [1402] "ENSG00000162543" "ENSG00000241635" "ENSG00000241635"
## [1405] "ENSG00000105668" "ENSG00000105698" "ENSG00000105698"
## [1408] "ENSG00000140455" "ENSG00000140455" "ENSG00000140455"
## [1411] "ENSG00000140455" "ENSG00000103404" "ENSG00000148429"
## [1414] "ENSG00000132952" "ENSG00000152818" "ENSG00000132612"
## [1417] "ENSG00000141252" "ENSG00000141252" "ENSG00000109072"
## [1420] "ENSG00000109072" "ENSG00000110799" "ENSG00000186153"
## [1423] "ENSG00000186153" "ENSG00000186153" "ENSG00000114127"
## [1426] "ENSG00000114127" "ENSG00000175155" "ENSG00000205189"
## [1429] "ENSG00000205189" "ENSG00000169155" "ENSG00000169155"
## [1432] "ENSG00000184828" "ENSG00000141664" "ENSG00000153786"
## [1435] "ENSG00000153786" "ENSG00000099904" "ENSG00000099904"
## [1438] "ENSG00000140836" "ENSG00000185650" "ENSG00000185650"
## [1441] "ENSG00000100711" "ENSG00000100711" "ENSG00000108175"
## [1444] "ENSG00000122515" "ENSG00000085644" "ENSG00000171940"
## [1447] "ENSG00000168813" "ENSG00000168813" "ENSG00000185252"
## [1450] "ENSG00000185252" "ENSG00000185252" "ENSG00000185252"
## [1453] "ENSG00000185252" "ENSG00000188372" "ENSG00000070476"
## [1456] "ENSG00000070476"# Convert the TSS to ensg as well
ref_gene_hg19_ensg <- merge(refGene_hg19_TSS, gene_id, by.x = c("V5"), by.y = c("Gene"))
all_ref_gene_hg19_ensg <- unique(ref_gene_hg19_ensg$ensg)
liver_ref_gene <- all_ref_gene_hg19_ensg %in% comb_kidney$ensg
# Revisions- run GO
# Merge ENSG with true/false
test_gene <- as.numeric(as.vector(liver_ref_gene))
names(test_gene) <- all_ref_gene_hg19_ensg
# Run topGO
go_data <- new("topGOdata",
ontology = "BP",
allGenes = test_gene,
geneSel = function(allScore){
return(allScore > 0)
},
nodeSize = 5,
annotationFun = annFUN.org,
mapping = "org.Hs.eg.db",
ID = "ensembl")##
## Building most specific GOs .....## ( 11450 GO terms found. )##
## Build GO DAG topology ..........## ( 15456 GO terms and 36153 relations. )##
## Annotating nodes ...............## ( 14505 genes annotated to the GO terms. )# Perform enrichment test
go_test <- runTest(go_data, algorithm = "weight01", statistic = "fisher")##
## -- Weight01 Algorithm --
##
## the algorithm is scoring 6020 nontrivial nodes
## parameters:
## test statistic: fisher##
## Level 18: 1 nodes to be scored (0 eliminated genes)##
## Level 17: 4 nodes to be scored (0 eliminated genes)##
## Level 16: 20 nodes to be scored (8 eliminated genes)##
## Level 15: 57 nodes to be scored (71 eliminated genes)##
## Level 14: 116 nodes to be scored (316 eliminated genes)##
## Level 13: 208 nodes to be scored (911 eliminated genes)##
## Level 12: 308 nodes to be scored (1979 eliminated genes)##
## Level 11: 502 nodes to be scored (4148 eliminated genes)##
## Level 10: 677 nodes to be scored (5660 eliminated genes)##
## Level 9: 819 nodes to be scored (7855 eliminated genes)##
## Level 8: 848 nodes to be scored (9971 eliminated genes)##
## Level 7: 873 nodes to be scored (11436 eliminated genes)##
## Level 6: 739 nodes to be scored (12657 eliminated genes)##
## Level 5: 470 nodes to be scored (13331 eliminated genes)##
## Level 4: 250 nodes to be scored (13846 eliminated genes)##
## Level 3: 105 nodes to be scored (14079 eliminated genes)##
## Level 2: 22 nodes to be scored (14207 eliminated genes)##
## Level 1: 1 nodes to be scored (14324 eliminated genes)go_table <- GenTable(go_data, weightFisher = go_test,
orderBy = "weightFisher", ranksOf = "weightFisher",
topNodes = sum(score(go_test) < .05))
go_table## GO.ID Term Annotated
## 1 GO:0030449 regulation of complement activation 41
## 2 GO:0042632 cholesterol homeostasis 68
## 3 GO:0006953 acute-phase response 42
## 4 GO:0006958 complement activation, classical pathway 28
## 5 GO:0070328 triglyceride homeostasis 29
## 6 GO:0009820 alkaloid metabolic process 5
## 7 GO:0002576 platelet degranulation 120
## 8 GO:0043691 reverse cholesterol transport 16
## 9 GO:0034384 high-density lipoprotein particle cleara... 11
## 10 GO:0006641 triglyceride metabolic process 90
## 11 GO:0055089 fatty acid homeostasis 12
## 12 GO:0046628 positive regulation of insulin receptor ... 13
## 13 GO:0006879 cellular iron ion homeostasis 57
## 14 GO:0006957 complement activation, alternative pathw... 14
## 15 GO:0034375 high-density lipoprotein particle remode... 15
## 16 GO:0008203 cholesterol metabolic process 121
## 17 GO:0090277 positive regulation of peptide hormone s... 79
## 18 GO:0006898 receptor-mediated endocytosis 264
## 19 GO:0008202 steroid metabolic process 265
## 20 GO:0006768 biotin metabolic process 10
## 21 GO:0034372 very-low-density lipoprotein particle re... 10
## 22 GO:0019835 cytolysis 35
## 23 GO:0007597 blood coagulation, intrinsic pathway 17
## 24 GO:1904179 positive regulation of adipose tissue de... 5
## 25 GO:0001907 killing by symbiont of host cells 5
## 26 GO:0006524 alanine catabolic process 5
## 27 GO:0042738 exogenous drug catabolic process 5
## 28 GO:0001889 liver development 124
## 29 GO:0007584 response to nutrient 168
## 30 GO:0010951 negative regulation of endopeptidase act... 213
## 31 GO:0017187 peptidyl-glutamic acid carboxylation 11
## 32 GO:0050892 intestinal absorption 34
## 33 GO:1904683 regulation of metalloendopeptidase activ... 6
## 34 GO:0015942 formate metabolic process 6
## 35 GO:0070141 response to UV-A 6
## 36 GO:0097267 omega-hydroxylase P450 pathway 6
## 37 GO:0006536 glutamate metabolic process 30
## 38 GO:0061138 morphogenesis of a branching epithelium 168
## 39 GO:0045944 positive regulation of transcription fro... 1010
## 40 GO:0034374 low-density lipoprotein particle remodel... 13
## 41 GO:0043433 negative regulation of DNA binding trans... 130
## 42 GO:0070301 cellular response to hydrogen peroxide 74
## 43 GO:0048538 thymus development 41
## 44 GO:0060059 embryonic retina morphogenesis in camera... 7
## 45 GO:0045620 negative regulation of lymphocyte differ... 40
## 46 GO:0045717 negative regulation of fatty acid biosyn... 14
## 47 GO:0002244 hematopoietic progenitor cell differenti... 155
## 48 GO:0045540 regulation of cholesterol biosynthetic p... 38
## 49 GO:0034380 high-density lipoprotein particle assemb... 15
## 50 GO:0045725 positive regulation of glycogen biosynth... 15
## 51 GO:0071276 cellular response to cadmium ion 24
## 52 GO:0046620 regulation of organ growth 89
## 53 GO:1903427 negative regulation of reactive oxygen s... 23
## 54 GO:0010918 positive regulation of mitochondrial mem... 8
## 55 GO:0015886 heme transport 8
## 56 GO:0051715 cytolysis in other organism 8
## 57 GO:0006548 histidine catabolic process 8
## 58 GO:0019216 regulation of lipid metabolic process 332
## 59 GO:0071559 response to transforming growth factor b... 203
## 60 GO:0019695 choline metabolic process 9
## 61 GO:0010984 regulation of lipoprotein particle clear... 17
## 62 GO:0090218 positive regulation of lipid kinase acti... 31
## 63 GO:0010894 negative regulation of steroid biosynthe... 22
## 64 GO:0046329 negative regulation of JNK cascade 30
## 65 GO:0006739 NADP metabolic process 33
## 66 GO:0016310 phosphorylation 2076
## 67 GO:0000098 sulfur amino acid catabolic process 9
## 68 GO:0042908 xenobiotic transport 9
## 69 GO:0070989 oxidative demethylation 9
## 70 GO:0030917 midbrain-hindbrain boundary development 9
## 71 GO:0046185 aldehyde catabolic process 9
## 72 GO:0070857 regulation of bile acid biosynthetic pro... 9
## 73 GO:0055091 phospholipid homeostasis 9
## 74 GO:0033344 cholesterol efflux 42
## 75 GO:0071157 negative regulation of cell cycle arrest 18
## 76 GO:0042593 glucose homeostasis 200
## 77 GO:0006869 lipid transport 311
## 78 GO:0051004 regulation of lipoprotein lipase activit... 18
## 79 GO:0050665 hydrogen peroxide biosynthetic process 10
## 80 GO:0051712 positive regulation of killing of cells ... 10
## 81 GO:0006853 carnitine shuttle 10
## 82 GO:1902237 positive regulation of endoplasmic retic... 10
## 83 GO:0001558 regulation of cell growth 356
## 84 GO:0050730 regulation of peptidyl-tyrosine phosphor... 210
## 85 GO:0042157 lipoprotein metabolic process 164
## 86 GO:0051081 nuclear envelope disassembly 43
## 87 GO:0019359 nicotinamide nucleotide biosynthetic pro... 27
## 88 GO:0034340 response to type I interferon 68
## 89 GO:0051647 nucleus localization 22
## 90 GO:0001867 complement activation, lectin pathway 11
## 91 GO:0019373 epoxygenase P450 pathway 11
## 92 GO:0071372 cellular response to follicle-stimulatin... 11
## 93 GO:0006559 L-phenylalanine catabolic process 11
## 94 GO:0045739 positive regulation of DNA repair 44
## 95 GO:0071560 cellular response to transforming growth... 200
## 96 GO:0006855 drug transmembrane transport 21
## 97 GO:0006368 transcription elongation from RNA polyme... 94
## 98 GO:0097421 liver regeneration 32
## 99 GO:0030514 negative regulation of BMP signaling pat... 44
## 100 GO:0006629 lipid metabolic process 1267
## 101 GO:0006520 cellular amino acid metabolic process 337
## 102 GO:0043903 regulation of symbiosis, encompassing mu... 197
## 103 GO:0042730 fibrinolysis 25
## 104 GO:0045830 positive regulation of isotype switching 22
## 105 GO:0099133 ATP hydrolysis coupled anion transmembra... 12
## 106 GO:1900119 positive regulation of execution phase o... 12
## 107 GO:2001241 positive regulation of extrinsic apoptot... 12
## 108 GO:0045721 negative regulation of gluconeogenesis 12
## 109 GO:0006695 cholesterol biosynthetic process 60
## 110 GO:0021915 neural tube development 153
## 111 GO:0010646 regulation of cell communication 3042
## 112 GO:0042177 negative regulation of protein catabolic... 102
## 113 GO:0033572 transferrin transport 34
## 114 GO:0022900 electron transport chain 143
## 115 GO:0042158 lipoprotein biosynthetic process 135
## 116 GO:2000646 positive regulation of receptor cataboli... 5
## 117 GO:0001887 selenium compound metabolic process 5
## 118 GO:0021894 cerebral cortex GABAergic interneuron de... 5
## 119 GO:1901523 icosanoid catabolic process 5
## 120 GO:0021814 cell motility involved in cerebral corte... 5
## 121 GO:0032430 positive regulation of phospholipase A2 ... 5
## 122 GO:0010991 negative regulation of SMAD protein comp... 5
## 123 GO:0019740 nitrogen utilization 5
## 124 GO:0060437 lung growth 5
## 125 GO:0048625 myoblast fate commitment 5
## 126 GO:0019626 short-chain fatty acid catabolic process 5
## 127 GO:1903753 negative regulation of p38MAPK cascade 5
## 128 GO:0010040 response to iron(II) ion 5
## 129 GO:0015911 plasma membrane long-chain fatty acid tr... 5
## 130 GO:0006572 tyrosine catabolic process 5
## 131 GO:0090324 negative regulation of oxidative phospho... 5
## 132 GO:0031999 negative regulation of fatty acid beta-o... 5
## 133 GO:0090118 receptor-mediated endocytosis involved i... 5
## 134 GO:0072049 comma-shaped body morphogenesis 5
## 135 GO:0010269 response to selenium ion 5
## 136 GO:0051642 centrosome localization 23
## 137 GO:0051147 regulation of muscle cell differentiatio... 179
## 138 GO:0034142 toll-like receptor 4 signaling pathway 28
## 139 GO:1900016 negative regulation of cytokine producti... 13
## 140 GO:0071397 cellular response to cholesterol 13
## 141 GO:0016540 protein autoprocessing 13
## 142 GO:0060712 spongiotrophoblast layer development 13
## 143 GO:0032436 positive regulation of proteasomal ubiqu... 75
## 144 GO:0055114 oxidation-reduction process 852
## 145 GO:0034504 protein localization to nucleus 341
## 146 GO:0006699 bile acid biosynthetic process 33
## 147 GO:0046718 viral entry into host cell 102
## 148 GO:2000463 positive regulation of excitatory postsy... 24
## 149 GO:0007411 axon guidance 219
## 150 GO:0042127 regulation of cell proliferation 1447
## 151 GO:0031325 positive regulation of cellular metaboli... 2748
## 152 GO:0002922 positive regulation of humoral immune re... 14
## 153 GO:0034356 NAD biosynthesis via nicotinamide ribosi... 14
## 154 GO:0042104 positive regulation of activated T cell ... 25
## 155 GO:0032270 positive regulation of cellular protein ... 1352
## 156 GO:0090335 regulation of brown fat cell differentia... 13
## 157 GO:0034763 negative regulation of transmembrane tra... 85
## 158 GO:0071156 regulation of cell cycle arrest 98
## 159 GO:0032375 negative regulation of cholesterol trans... 14
## 160 GO:2000659 regulation of interleukin-1-mediated sig... 6
## 161 GO:1990535 neuron projection maintenance 6
## 162 GO:1902946 protein localization to early endosome 6
## 163 GO:0042940 D-amino acid transport 6
## 164 GO:0033860 regulation of NAD(P)H oxidase activity 6
## 165 GO:1900107 regulation of nodal signaling pathway 6
## 166 GO:0046498 S-adenosylhomocysteine metabolic process 6
## 167 GO:1905049 negative regulation of metallopeptidase ... 6
## 168 GO:1903896 positive regulation of IRE1-mediated unf... 6
## 169 GO:1902459 positive regulation of stem cell populat... 6
## 170 GO:0042426 choline catabolic process 6
## 171 GO:0031659 positive regulation of cyclin-dependent ... 6
## 172 GO:0034382 chylomicron remnant clearance 6
## 173 GO:2000343 positive regulation of chemokine (C-X-C ... 6
## 174 GO:0006545 glycine biosynthetic process 6
## 175 GO:0010793 regulation of mRNA export from nucleus 6
## 176 GO:1905668 positive regulation of protein localizat... 6
## 177 GO:1903364 positive regulation of cellular protein ... 186
## 178 GO:0021879 forebrain neuron differentiation 49
## 179 GO:0019432 triglyceride biosynthetic process 31
## 180 GO:0006461 protein complex assembly 1257
## 181 GO:0021542 dentate gyrus development 15
## 182 GO:0090201 negative regulation of release of cytoch... 15
## 183 GO:0000060 protein import into nucleus, translocati... 45
## 184 GO:0046326 positive regulation of glucose import 39
## 185 GO:0035774 positive regulation of insulin secretion... 26
## 186 GO:0007628 adult walking behavior 26
## 187 GO:0032088 negative regulation of NF-kappaB transcr... 66
## 188 GO:0008152 metabolic process 9852
## 189 GO:0048013 ephrin receptor signaling pathway 81
## 190 GO:0043392 negative regulation of DNA binding 47
## 191 GO:0009887 animal organ morphogenesis 919
## 192 GO:0032094 response to food 34
## 193 GO:0021756 striatum development 16
## 194 GO:0060644 mammary gland epithelial cell differenti... 16
## 195 GO:1903830 magnesium ion transmembrane transport 16
## 196 GO:0007595 lactation 40
## 197 GO:0000122 negative regulation of transcription fro... 727
## 198 GO:0097190 apoptotic signaling pathway 543
## 199 GO:0031663 lipopolysaccharide-mediated signaling pa... 48
## 200 GO:0030178 negative regulation of Wnt signaling pat... 186
## 201 GO:0030195 negative regulation of blood coagulation 46
## 202 GO:0019674 NAD metabolic process 64
## 203 GO:1902373 negative regulation of mRNA catabolic pr... 40
## 204 GO:0032786 positive regulation of DNA-templated tra... 20
## 205 GO:0033598 mammary gland epithelial cell proliferat... 22
## 206 GO:0034638 phosphatidylcholine catabolic process 7
## 207 GO:0010898 positive regulation of triglyceride cata... 7
## 208 GO:0070544 histone H3-K36 demethylation 7
## 209 GO:0010886 positive regulation of cholesterol stora... 7
## 210 GO:0060052 neurofilament cytoskeleton organization 7
## 211 GO:0021830 interneuron migration from the subpalliu... 7
## 212 GO:1901679 nucleotide transmembrane transport 7
## 213 GO:0000042 protein targeting to Golgi 7
## 214 GO:0051045 negative regulation of membrane protein ... 7
## 215 GO:1904468 negative regulation of tumor necrosis fa... 7
## 216 GO:0044332 Wnt signaling pathway involved in dorsal... 7
## 217 GO:0035356 cellular triglyceride homeostasis 7
## 218 GO:0032287 peripheral nervous system myelin mainten... 7
## 219 GO:0060463 lung lobe morphogenesis 7
## 220 GO:0046439 L-cysteine metabolic process 7
## 221 GO:1903874 ferrous iron transmembrane transport 7
## 222 GO:0031666 positive regulation of lipopolysaccharid... 7
## 223 GO:0042167 heme catabolic process 7
## 224 GO:2000192 negative regulation of fatty acid transp... 7
## 225 GO:0090336 positive regulation of brown fat cell di... 7
## 226 GO:0042758 long-chain fatty acid catabolic process 7
## 227 GO:0030300 regulation of intestinal cholesterol abs... 7
## 228 GO:0006600 creatine metabolic process 7
## 229 GO:2000271 positive regulation of fibroblast apopto... 7
## 230 GO:0008090 retrograde axonal transport 7
## 231 GO:0002679 respiratory burst involved in defense re... 7
## 232 GO:0032071 regulation of endodeoxyribonuclease acti... 7
## 233 GO:0043627 response to estrogen 66
## 234 GO:0051346 negative regulation of hydrolase activit... 362
## 235 GO:0061045 negative regulation of wound healing 63
## 236 GO:0017144 drug metabolic process 24
## 237 GO:0007041 lysosomal transport 86
## 238 GO:0001938 positive regulation of endothelial cell ... 70
## 239 GO:0051000 positive regulation of nitric-oxide synt... 17
## 240 GO:1902176 negative regulation of oxidative stress-... 17
## 241 GO:0031100 animal organ regeneration 74
## 242 GO:0032869 cellular response to insulin stimulus 176
## 243 GO:0032007 negative regulation of TOR signaling 40
## 244 GO:0018279 protein N-linked glycosylation via aspar... 42
## 245 GO:0045599 negative regulation of fat cell differen... 42
## 246 GO:0032148 activation of protein kinase B activity 29
## 247 GO:0001503 ossification 343
## 248 GO:1902680 positive regulation of RNA biosynthetic ... 1299
## 249 GO:0030324 lung development 164
## 250 GO:0060021 palate development 87
## 251 GO:2000257 regulation of protein activation cascade 42
## 252 GO:0042493 response to drug 373
## 253 GO:0001568 blood vessel development 570
## 254 GO:0031016 pancreas development 67
## 255 GO:0006778 porphyrin-containing compound metabolic ... 34
## 256 GO:1900076 regulation of cellular response to insul... 60
## 257 GO:0090239 regulation of histone H4 acetylation 10
## 258 GO:0019627 urea metabolic process 11
## 259 GO:0015865 purine nucleotide transport 12
## 260 GO:0061620 glycolytic process through glucose-6-pho... 24
## 261 GO:0050820 positive regulation of coagulation 24
## 262 GO:0006476 protein deacetylation 92
## 263 GO:0046631 alpha-beta T cell activation 114
## 264 GO:0098902 regulation of membrane depolarization du... 6
## 265 GO:1902959 regulation of aspartic-type endopeptidas... 6
## 266 GO:0052433 modulation by organism of apoptotic proc... 6
## 267 GO:0070943 neutrophil mediated killing of symbiont ... 6
## 268 GO:0046884 follicle-stimulating hormone secretion 6
## 269 GO:0052040 modulation by symbiont of host programme... 6
## 270 GO:0051340 regulation of ligase activity 7
## 271 GO:0035733 hepatic stellate cell activation 7
## 272 GO:0046838 phosphorylated carbohydrate dephosphoryl... 10
## 273 GO:1901160 primary amino compound metabolic process 13
## 274 GO:0036475 neuron death in response to oxidative st... 21
## 275 GO:0042773 ATP synthesis coupled electron transport 66
## 276 GO:0010039 response to iron ion 33
## 277 GO:0006505 GPI anchor metabolic process 28
## 278 GO:1900182 positive regulation of protein localizat... 124
## 279 GO:0052652 cyclic purine nucleotide metabolic proce... 145
## 280 GO:0035909 aorta morphogenesis 30
## 281 GO:0014911 positive regulation of smooth muscle cel... 30
## 282 GO:0035435 phosphate ion transmembrane transport 18
## 283 GO:0006465 signal peptide processing 18
## 284 GO:0061003 positive regulation of dendritic spine m... 18
## 285 GO:0035264 multicellular organism growth 137
## 286 GO:0014068 positive regulation of phosphatidylinosi... 58
## 287 GO:0043405 regulation of MAP kinase activity 303
## 288 GO:0008595 anterior/posterior axis specification, e... 13
## 289 GO:0050746 regulation of lipoprotein metabolic proc... 13
## 290 GO:0045657 positive regulation of monocyte differen... 8
## 291 GO:0010873 positive regulation of cholesterol ester... 8
## 292 GO:0001561 fatty acid alpha-oxidation 8
## 293 GO:0071073 positive regulation of phospholipid bios... 8
## 294 GO:1900364 negative regulation of mRNA polyadenylat... 8
## 295 GO:1900112 regulation of histone H3-K9 trimethylati... 8
## 296 GO:0008635 activation of cysteine-type endopeptidas... 8
## 297 GO:0007262 STAT protein import into nucleus 8
## 298 GO:0006390 transcription from mitochondrial promote... 8
## 299 GO:0015866 ADP transport 8
## 300 GO:0046487 glyoxylate metabolic process 8
## 301 GO:0051573 negative regulation of histone H3-K9 met... 8
## 302 GO:0032926 negative regulation of activin receptor ... 8
## 303 GO:0032933 SREBP signaling pathway 8
## 304 GO:0006591 ornithine metabolic process 8
## 305 GO:1990314 cellular response to insulin-like growth... 8
## 306 GO:0031937 positive regulation of chromatin silenci... 8
## 307 GO:0090400 stress-induced premature senescence 8
## 308 GO:0051897 positive regulation of protein kinase B ... 137
## Significant Expected weightFisher
## 1 13 1.87 1.8e-08
## 2 15 3.10 1.4e-07
## 3 11 1.91 6.7e-07
## 4 9 1.28 2.5e-06
## 5 10 1.32 3.2e-06
## 6 4 0.23 2.1e-05
## 7 17 5.47 3.0e-05
## 8 6 0.73 4.7e-05
## 9 5 0.50 7.1e-05
## 10 16 4.10 8.7e-05
## 11 5 0.55 0.00012
## 12 5 0.59 0.00018
## 13 10 2.60 0.00022
## 14 5 0.64 0.00027
## 15 5 0.68 0.00040
## 16 21 5.51 0.00041
## 17 12 3.60 0.00044
## 18 25 12.03 0.00044
## 19 41 12.08 0.00047
## 20 4 0.46 0.00072
## 21 4 0.46 0.00072
## 22 9 1.59 0.00074
## 23 5 0.77 0.00076
## 24 3 0.23 0.00088
## 25 3 0.23 0.00088
## 26 3 0.23 0.00088
## 27 3 0.23 0.00088
## 28 17 5.65 0.00091
## 29 21 7.66 0.00100
## 30 18 9.71 0.00103
## 31 4 0.50 0.00109
## 32 8 1.55 0.00155
## 33 3 0.27 0.00170
## 34 3 0.27 0.00170
## 35 3 0.27 0.00170
## 36 3 0.27 0.00170
## 37 6 1.37 0.00204
## 38 9 7.66 0.00208
## 39 67 46.03 0.00213
## 40 4 0.59 0.00220
## 41 16 5.92 0.00225
## 42 9 3.37 0.00227
## 43 7 1.87 0.00230
## 44 3 0.32 0.00287
## 45 4 1.82 0.00288
## 46 4 0.64 0.00297
## 47 12 7.06 0.00328
## 48 7 1.73 0.00336
## 49 4 0.68 0.00390
## 50 4 0.68 0.00390
## 51 5 1.09 0.00400
## 52 8 4.06 0.00442
## 53 4 1.05 0.00444
## 54 3 0.36 0.00444
## 55 3 0.36 0.00444
## 56 3 0.36 0.00444
## 57 3 0.36 0.00444
## 58 39 15.13 0.00523
## 59 15 9.25 0.00596
## 60 4 0.41 0.00600
## 61 4 0.77 0.00601
## 62 3 1.41 0.00604
## 63 5 1.00 0.00630
## 64 5 1.37 0.00631
## 65 5 1.50 0.00632
## 66 116 94.60 0.00636
## 67 3 0.41 0.00644
## 68 3 0.41 0.00644
## 69 3 0.41 0.00644
## 70 3 0.41 0.00644
## 71 3 0.41 0.00644
## 72 3 0.41 0.00644
## 73 3 0.41 0.00644
## 74 8 1.91 0.00772
## 75 4 0.82 0.00784
## 76 20 9.11 0.00840
## 77 38 14.17 0.00841
## 78 4 0.82 0.00887
## 79 3 0.46 0.00889
## 80 3 0.46 0.00889
## 81 3 0.46 0.00889
## 82 3 0.46 0.00889
## 83 28 16.22 0.00937
## 84 18 9.57 0.01152
## 85 14 7.47 0.01162
## 86 6 1.96 0.01162
## 87 5 1.23 0.01163
## 88 6 3.10 0.01166
## 89 3 1.00 0.01169
## 90 3 0.50 0.01181
## 91 3 0.50 0.01181
## 92 3 0.50 0.01181
## 93 3 0.50 0.01181
## 94 6 2.01 0.01226
## 95 13 9.11 0.01269
## 96 4 0.96 0.01377
## 97 10 4.28 0.01386
## 98 5 1.46 0.01404
## 99 6 2.01 0.01413
## 100 99 57.74 0.01441
## 101 36 15.36 0.01480
## 102 17 8.98 0.01488
## 103 5 1.14 0.01514
## 104 4 1.00 0.01519
## 105 3 0.55 0.01522
## 106 3 0.55 0.01522
## 107 3 0.55 0.01522
## 108 3 0.55 0.01522
## 109 11 2.73 0.01581
## 110 13 6.97 0.01599
## 111 165 138.63 0.01725
## 112 11 4.65 0.01770
## 113 5 1.55 0.01801
## 114 12 6.52 0.01863
## 115 8 6.15 0.01891
## 116 2 0.23 0.01892
## 117 2 0.23 0.01892
## 118 2 0.23 0.01892
## 119 2 0.23 0.01892
## 120 2 0.23 0.01892
## 121 2 0.23 0.01892
## 122 2 0.23 0.01892
## 123 2 0.23 0.01892
## 124 2 0.23 0.01892
## 125 2 0.23 0.01892
## 126 2 0.23 0.01892
## 127 2 0.23 0.01892
## 128 2 0.23 0.01892
## 129 2 0.23 0.01892
## 130 2 0.23 0.01892
## 131 2 0.23 0.01892
## 132 2 0.23 0.01892
## 133 2 0.23 0.01892
## 134 2 0.23 0.01892
## 135 2 0.23 0.01892
## 136 4 1.05 0.01896
## 137 9 8.16 0.01897
## 138 4 1.28 0.01911
## 139 3 0.59 0.01913
## 140 3 0.59 0.01913
## 141 3 0.59 0.01913
## 142 3 0.59 0.01913
## 143 8 3.42 0.02051
## 144 59 38.83 0.02063
## 145 27 15.54 0.02102
## 146 7 1.50 0.02168
## 147 11 4.65 0.02172
## 148 4 1.09 0.02196
## 149 17 9.98 0.02232
## 150 79 65.94 0.02293
## 151 172 125.23 0.02332
## 152 3 0.64 0.02354
## 153 3 0.64 0.02354
## 154 4 1.14 0.02522
## 155 81 61.61 0.02602
## 156 4 0.59 0.02739
## 157 7 3.87 0.02741
## 158 7 4.47 0.02746
## 159 3 0.64 0.02747
## 160 2 0.27 0.02752
## 161 2 0.27 0.02752
## 162 2 0.27 0.02752
## 163 2 0.27 0.02752
## 164 2 0.27 0.02752
## 165 2 0.27 0.02752
## 166 2 0.27 0.02752
## 167 2 0.27 0.02752
## 168 2 0.27 0.02752
## 169 2 0.27 0.02752
## 170 2 0.27 0.02752
## 171 2 0.27 0.02752
## 172 2 0.27 0.02752
## 173 2 0.27 0.02752
## 174 2 0.27 0.02752
## 175 2 0.27 0.02752
## 176 2 0.27 0.02752
## 177 13 8.48 0.02820
## 178 6 2.23 0.02829
## 179 5 1.41 0.02831
## 180 73 57.28 0.02845
## 181 3 0.68 0.02845
## 182 3 0.68 0.02845
## 183 6 2.05 0.02861
## 184 5 1.78 0.02871
## 185 4 1.18 0.02877
## 186 4 1.18 0.02877
## 187 7 3.01 0.03008
## 188 492 448.96 0.03079
## 189 8 3.69 0.03092
## 190 6 2.14 0.03242
## 191 51 41.88 0.03332
## 192 4 1.55 0.03384
## 193 3 0.73 0.03386
## 194 3 0.73 0.03386
## 195 3 0.73 0.03386
## 196 5 1.82 0.03412
## 197 45 33.13 0.03431
## 198 39 24.74 0.03565
## 199 7 2.19 0.03633
## 200 11 8.48 0.03673
## 201 7 2.10 0.03705
## 202 7 2.92 0.03713
## 203 4 1.82 0.03733
## 204 3 0.91 0.03734
## 205 3 1.00 0.03735
## 206 2 0.32 0.03738
## 207 2 0.32 0.03738
## 208 2 0.32 0.03738
## 209 2 0.32 0.03738
## 210 2 0.32 0.03738
## 211 2 0.32 0.03738
## 212 2 0.32 0.03738
## 213 2 0.32 0.03738
## 214 2 0.32 0.03738
## 215 2 0.32 0.03738
## 216 2 0.32 0.03738
## 217 2 0.32 0.03738
## 218 2 0.32 0.03738
## 219 2 0.32 0.03738
## 220 2 0.32 0.03738
## 221 2 0.32 0.03738
## 222 2 0.32 0.03738
## 223 2 0.32 0.03738
## 224 2 0.32 0.03738
## 225 2 0.32 0.03738
## 226 2 0.32 0.03738
## 227 2 0.32 0.03738
## 228 2 0.32 0.03738
## 229 2 0.32 0.03738
## 230 2 0.32 0.03738
## 231 2 0.32 0.03738
## 232 2 0.32 0.03738
## 233 8 3.01 0.03791
## 234 26 16.50 0.03864
## 235 10 2.87 0.03902
## 236 7 1.09 0.03920
## 237 8 3.92 0.03948
## 238 7 3.19 0.03971
## 239 3 0.77 0.03976
## 240 3 0.77 0.03976
## 241 10 3.37 0.04019
## 242 15 8.02 0.04037
## 243 6 1.82 0.04080
## 244 5 1.91 0.04099
## 245 5 1.91 0.04099
## 246 4 1.32 0.04109
## 247 19 15.63 0.04129
## 248 85 59.20 0.04369
## 249 12 7.47 0.04431
## 250 8 3.96 0.04448
## 251 14 1.91 0.04480
## 252 32 17.00 0.04486
## 253 31 25.98 0.04523
## 254 8 3.05 0.04527
## 255 6 1.55 0.04533
## 256 6 2.73 0.04541
## 257 3 0.46 0.04546
## 258 3 0.50 0.04546
## 259 3 0.55 0.04547
## 260 3 1.09 0.04550
## 261 3 1.09 0.04550
## 262 6 4.19 0.04551
## 263 7 5.20 0.04551
## 264 2 0.27 0.04552
## 265 2 0.27 0.04552
## 266 2 0.27 0.04552
## 267 2 0.27 0.04552
## 268 2 0.27 0.04552
## 269 2 0.27 0.04552
## 270 2 0.32 0.04552
## 271 2 0.32 0.04552
## 272 2 0.46 0.04553
## 273 2 0.59 0.04554
## 274 2 0.96 0.04556
## 275 4 3.01 0.04557
## 276 7 1.50 0.04557
## 277 2 1.28 0.04559
## 278 9 5.65 0.04561
## 279 6 6.61 0.04568
## 280 4 1.37 0.04576
## 281 4 1.37 0.04607
## 282 3 0.82 0.04615
## 283 3 0.82 0.04615
## 284 3 0.82 0.04615
## 285 13 6.24 0.04712
## 286 6 2.64 0.04773
## 287 19 13.81 0.04788
## 288 3 0.59 0.04826
## 289 3 0.59 0.04826
## 290 2 0.36 0.04837
## 291 2 0.36 0.04837
## 292 2 0.36 0.04837
## 293 2 0.36 0.04837
## 294 2 0.36 0.04837
## 295 2 0.36 0.04837
## 296 2 0.36 0.04837
## 297 2 0.36 0.04837
## 298 2 0.36 0.04837
## 299 2 0.36 0.04837
## 300 2 0.36 0.04837
## 301 2 0.36 0.04837
## 302 2 0.36 0.04837
## 303 2 0.36 0.04837
## 304 2 0.36 0.04837
## 305 2 0.36 0.04837
## 306 2 0.36 0.04837
## 307 2 0.36 0.04837
## 308 11 6.24 0.04848 go_table_liver <- as.data.frame(cbind(go_table$GO.ID, go_table$weightFisher))
write.table(go_table_liver, "../data/go_table_liver.txt", quote = FALSE, row.names = FALSE, col.names = FALSE)Lung
# Find the heart hypo
heart_hypo <- human_chimp_heart_specific_25_conserved_H3K27_LUNG[which(human_chimp_heart_specific_25_conserved_H3K27_LUNG$V4 == "hyper" & human_chimp_heart_specific_25_conserved_H3K27_LUNG$V6 != "-1"),]
heart_hypo <- heart_hypo[complete.cases(heart_hypo), ]
# Find the closest gene
closest_heart <- bedr(input = list(a = heart_hypo[,1:3], b = refGene_hg19_TSS), method = "closest", check.chr = FALSE)## * Processing input (1): a
## CONVERT TO BED
## * Checking input type... PASS
## Input seems to be in bed format but chr/start/end column names are missing
## VALIDATE REGIONS
## * Check if index is a string... PASS
## * Check index pattern... PASS
## * Check for missing values... PASS
## * Check for larger start position... PASS.
## * Check if zero based... PASS
## * Checking sort order... PASS
## * Checking for overlapping 'contiguous' regions... PASS
## * Processing input (2): b
## CONVERT TO BED
## * Checking input type... PASS
## Input seems to be in bed format but chr/start/end column names are missing
## VALIDATE REGIONS
## * Check if index is a string... PASS
## * Check index pattern... PASS
## * Check for missing values... PASS
## * Check for larger start position... PASS.
## * Check if zero based... PASS
## * Checking sort order... FAIL
## The input for object is not *lexographically* ordered!
## This can cause unexpected results for some set operations.
## try: x <- bedr.sort.region(x)
## * Checking for overlapping 'contiguous' regions... FAIL
## The input for object has overlapping features!
## This can cause unexpected results for some set operations.
## i.e. x <- bedr.merge.region(x)
## bedtools closest -a /var/folders/rf/qrcw6ncj05z1pc_pq9xzw3540000gn/T//RtmpXK3I56/a_bc552850ec24.bed -b /var/folders/rf/qrcw6ncj05z1pc_pq9xzw3540000gn/T//RtmpXK3I56/b_bc557772e5c1.bed# Convert the gene name to ensg
gene_id <- read.table("../../../Reg_Evo_Primates/data/ENSG_GENE_HG19.csv", stringsAsFactors = FALSE, header=TRUE, sep = ",")
comb_kidney <- merge(closest_heart, gene_id, by.x = c("V8"), by.y = c("Gene"))
comb_kidney$ensg## [1] "ENSG00000177674" "ENSG00000177674" "ENSG00000177674"
## [4] "ENSG00000177674" "ENSG00000177674" "ENSG00000130762"
## [7] "ENSG00000117411" "ENSG00000026508" "ENSG00000026508"
## [10] "ENSG00000026508" "ENSG00000026508" "ENSG00000026508"
## [13] "ENSG00000026508" "ENSG00000026508" "ENSG00000026508"
## [16] "ENSG00000114861" "ENSG00000114315" "ENSG00000131149"
## [19] "ENSG00000198951" "ENSG00000196498" "ENSG00000196498"
## [22] "ENSG00000196498" "ENSG00000170322" "ENSG00000100100"
## [25] "ENSG00000100100" "ENSG00000185917" "ENSG00000185917"
## [28] "ENSG00000167323" "ENSG00000143643" "ENSG00000143643"
## [31] "ENSG00000153786" "ENSG00000153786" "ENSG00000171940"
## [34] "ENSG00000183621" "ENSG00000183621" "ENSG00000183621"
## [37] "ENSG00000183621" "ENSG00000183621" "ENSG00000183621"
## [40] "ENSG00000183621"# Convert the TSS to ensg as well
ref_gene_hg19_ensg <- merge(refGene_hg19_TSS, gene_id, by.x = c("V5"), by.y = c("Gene"))
all_ref_gene_hg19_ensg <- unique(ref_gene_hg19_ensg$ensg)
heart_ref_gene <- all_ref_gene_hg19_ensg %in% comb_kidney$ensg
# Revisions- run GO
# Merge ENSG with true/false
test_gene <- as.numeric(as.vector(heart_ref_gene))
names(test_gene) <- all_ref_gene_hg19_ensg
# Run topGO
go_data <- new("topGOdata",
ontology = "BP",
allGenes = test_gene,
geneSel = function(allScore){
return(allScore > 0)
},
nodeSize = 5,
annotationFun = annFUN.org,
mapping = "org.Hs.eg.db",
ID = "ensembl")##
## Building most specific GOs .....## ( 11450 GO terms found. )##
## Build GO DAG topology ..........## ( 15456 GO terms and 36153 relations. )##
## Annotating nodes ...............## ( 14505 genes annotated to the GO terms. )# Perform enrichment test
go_test <- runTest(go_data, algorithm = "weight01", statistic = "fisher")##
## -- Weight01 Algorithm --
##
## the algorithm is scoring 1110 nontrivial nodes
## parameters:
## test statistic: fisher##
## Level 15: 3 nodes to be scored (0 eliminated genes)##
## Level 14: 5 nodes to be scored (0 eliminated genes)##
## Level 13: 23 nodes to be scored (86 eliminated genes)##
## Level 12: 44 nodes to be scored (192 eliminated genes)##
## Level 11: 76 nodes to be scored (1997 eliminated genes)##
## Level 10: 103 nodes to be scored (3134 eliminated genes)##
## Level 9: 112 nodes to be scored (5198 eliminated genes)##
## Level 8: 124 nodes to be scored (6736 eliminated genes)##
## Level 7: 155 nodes to be scored (7719 eliminated genes)##
## Level 6: 166 nodes to be scored (10145 eliminated genes)##
## Level 5: 144 nodes to be scored (11508 eliminated genes)##
## Level 4: 90 nodes to be scored (13129 eliminated genes)##
## Level 3: 48 nodes to be scored (13788 eliminated genes)##
## Level 2: 16 nodes to be scored (14144 eliminated genes)##
## Level 1: 1 nodes to be scored (14277 eliminated genes)go_table <- GenTable(go_data, weightFisher = go_test,
orderBy = "weightFisher", ranksOf = "weightFisher",
topNodes = sum(score(go_test) < .01))
go_table## GO.ID Term Annotated
## 1 GO:0035019 somatic stem cell population maintenance 61
## 2 GO:0061009 common bile duct development 5
## 3 GO:0032237 activation of store-operated calcium cha... 5
## 4 GO:0072049 comma-shaped body morphogenesis 5
## 5 GO:0003310 pancreatic A cell differentiation 5
## 6 GO:0009912 auditory receptor cell fate commitment 6
## 7 GO:1901533 negative regulation of hematopoietic pro... 6
## 8 GO:2001182 regulation of interleukin-12 secretion 6
## 9 GO:2000973 regulation of pro-B cell differentiation 6
## 10 GO:0021861 forebrain radial glial cell differentiat... 6
## 11 GO:0018230 peptidyl-L-cysteine S-palmitoylation 6
## 12 GO:1903039 positive regulation of leukocyte cell-ce... 195
## 13 GO:0046477 glycosylceramide catabolic process 7
## 14 GO:1903799 negative regulation of production of miR... 7
## 15 GO:0061101 neuroendocrine cell differentiation 7
## 16 GO:0072050 S-shaped body morphogenesis 7
## 17 GO:0061626 pharyngeal arch artery morphogenesis 7
## 18 GO:0042117 monocyte activation 7
## 19 GO:0048664 neuron fate determination 7
## 20 GO:0045607 regulation of auditory receptor cell dif... 8
## 21 GO:0072282 metanephric nephron tubule morphogenesis 8
## 22 GO:0097084 vascular smooth muscle cell development 8
## 23 GO:0007262 STAT protein import into nucleus 8
## 24 GO:0016139 glycoside catabolic process 8
## 25 GO:1903726 negative regulation of phospholipid meta... 8
## 26 GO:0061309 cardiac neural crest cell development in... 9
## 27 GO:0018026 peptidyl-lysine monomethylation 9
## 28 GO:0003266 regulation of secondary heart field card... 9
## 29 GO:0030917 midbrain-hindbrain boundary development 9
## 30 GO:0070486 leukocyte aggregation 9
## 31 GO:0072567 chemokine (C-X-C motif) ligand 2 product... 9
## Significant Expected weightFisher
## 1 2 0.06 0.0018
## 2 1 0.01 0.0052
## 3 1 0.01 0.0052
## 4 1 0.01 0.0052
## 5 1 0.01 0.0052
## 6 1 0.01 0.0062
## 7 1 0.01 0.0062
## 8 1 0.01 0.0062
## 9 1 0.01 0.0062
## 10 1 0.01 0.0062
## 11 1 0.01 0.0062
## 12 2 0.20 0.0068
## 13 1 0.01 0.0072
## 14 1 0.01 0.0072
## 15 1 0.01 0.0072
## 16 1 0.01 0.0072
## 17 1 0.01 0.0072
## 18 1 0.01 0.0072
## 19 1 0.01 0.0072
## 20 1 0.01 0.0082
## 21 1 0.01 0.0082
## 22 1 0.01 0.0082
## 23 1 0.01 0.0082
## 24 1 0.01 0.0082
## 25 1 0.01 0.0082
## 26 1 0.01 0.0093
## 27 1 0.01 0.0093
## 28 1 0.01 0.0093
## 29 1 0.01 0.0093
## 30 1 0.01 0.0093
## 31 1 0.01 0.0093Run clusterProfiler
#head(heart_ref_gene)
#length(heart_ref_gene)
#head(liver_ref_gene)
#length(liver_ref_gene)
#mydf <- as.data.frame(cbind(all_ref_gene_hg19_ensg, heart_ref_gene, liver_ref_gene))
#colnames(mydf) <- c("ensg", "heart", "liver")
#xx.formula.twogroups <- compareCluster(mydf$heart, data = mydf, fun="enrichGO", universe = df$ensg,
# OrgDb = org.Hs.eg.db)
#dotplot(xx.formula.twogroups, showCategory=5, includeAll=FALSE)## Reivigo
# If you don't have the ggplot2 package installed, uncomment the following line:
# install.packages( "ggplot2" );
library( ggplot2 );
# --------------------------------------------------------------------------
# If you don't have the scales package installed, uncomment the following line:
# install.packages( "scales" );
library( scales );## Warning: package 'scales' was built under R version 3.4.4library(ggrepel)
# --------------------------------------------------------------------------
# Here is your data from REVIGO. Scroll down for plot configuration options.
revigo.names <- c("term_ID","description","frequency_%","plot_X","plot_Y","plot_size","value","uniqueness","dispensability");
revigo.data <- rbind(c("GO:0009113","purine nucleobase biosynthetic process", 0.124,-4.742,-3.192, 4.202, 0.0005,0.769,0.000),
c("GO:0033197","response to vitamin E", 0.000, 7.179,-3.273, 1.740, 0.0005,0.849,0.000),
c("GO:0019043","establishment of viral latency", 0.001,-5.135, 5.016, 1.959, 0.0187,0.945,0.018),
c("GO:0006501","C-terminal protein lipidation", 0.004, 0.003,-5.207, 2.693, 0.0120,0.913,0.100),
c("GO:0060509","Type I pneumocyte differentiation", 0.001,-0.420, 6.928, 2.121, 0.0134,0.698,0.103),
c("GO:1903078","positive regulation of protein localization to plasma membrane", 0.010, 1.621, 0.720, 3.123, 0.0068,0.688,0.118),
c("GO:0033632","regulation of cell-cell adhesion mediated by integrin", 0.002, 2.909, 0.449, 2.378, 0.0160,0.835,0.132),
c("GO:2000109","regulation of macrophage apoptotic process", 0.001, 2.079,-2.113, 2.188, 0.0187,0.768,0.132),
c("GO:0032007","negative regulation of TOR signaling", 0.016, 5.974, 0.117, 3.325, 0.0052,0.640,0.175),
c("GO:0043547","positive regulation of GTPase activity", 0.470, 4.687, 1.488, 4.780, 0.0054,0.838,0.179),
c("GO:0009256","10-formyltetrahydrofolate metabolic process", 0.002,-6.344,-1.530, 2.474, 0.0134,0.809,0.185),
c("GO:0070141","response to UV-A", 0.002, 6.586,-3.433, 2.369, 0.0160,0.883,0.188),
c("GO:0071499","cellular response to laminar fluid shear stress", 0.001, 6.586,-2.544, 2.143, 0.0187,0.852,0.211),
c("GO:0006139","nucleobase-containing compound metabolic process",26.547,-4.434,-2.184, 6.532, 0.0499,0.889,0.214),
c("GO:0018242","protein O-linked glycosylation via serine", 0.001,-1.029,-5.988, 2.117, 0.0160,0.818,0.218),
c("GO:0015942","formate metabolic process", 0.065,-5.564,-1.813, 3.921, 0.0160,0.815,0.246),
c("GO:0070601","centromeric sister chromatid cohesion", 0.004,-1.575,-1.033, 2.749, 0.0266,0.829,0.253),
c("GO:0006863","purine nucleobase transport", 0.002,-4.632, 3.161, 2.464, 0.0160,0.877,0.268),
c("GO:0032026","response to magnesium ion", 0.002, 6.932,-3.864, 2.389, 0.0473,0.863,0.290),
c("GO:0070375","ERK5 cascade", 0.001, 3.865,-3.823, 2.041, 0.0134,0.729,0.303),
c("GO:0060070","canonical Wnt signaling pathway", 0.061, 4.991,-1.442, 3.893, 0.0066,0.678,0.315),
c("GO:0032486","Rap protein signal transduction", 0.002, 6.347,-0.924, 2.458, 0.0344,0.763,0.318),
c("GO:0061430","bone trabecula morphogenesis", 0.003,-2.302, 6.815, 2.553, 0.0344,0.798,0.326),
c("GO:0001957","intramembranous ossification", 0.001, 0.332, 7.419, 2.167, 0.0160,0.766,0.343),
c("GO:0009120","deoxyribonucleoside metabolic process", 0.006,-4.177,-4.617, 2.908, 0.0187,0.800,0.350),
c("GO:0007386","compartment pattern specification", 0.001, 0.048, 7.245, 2.111, 0.0134,0.749,0.356),
c("GO:0045351","type I interferon biosynthetic process", 0.002, 0.347, 6.406, 2.505, 0.0318,0.743,0.357),
c("GO:0001821","histamine secretion", 0.001,-3.314, 2.812, 2.143, 0.0240,0.877,0.358),
c("GO:0042321","negative regulation of circadian sleep/wake cycle, sleep", 0.001, 2.446, 5.266, 1.845, 0.0134,0.663,0.369),
c("GO:0006744","ubiquinone biosynthetic process", 0.136,-5.223,-2.418, 4.242, 0.0396,0.786,0.389),
c("GO:0009146","purine nucleoside triphosphate catabolic process", 0.035,-3.242,-4.729, 3.647, 0.0134,0.749,0.391),
c("GO:0046085","adenosine metabolic process", 0.003,-4.584,-4.405, 2.524, 0.0266,0.804,0.409),
c("GO:0060405","regulation of penile erection", 0.000, 1.798, 6.090, 1.716, 0.0160,0.701,0.410),
c("GO:0045725","positive regulation of glycogen biosynthetic process", 0.003, 0.849,-3.136, 2.547, 0.0396,0.723,0.414),
c("GO:1901642","nucleoside transmembrane transport", 0.047,-3.483, 2.842, 3.781, 0.0187,0.860,0.432),
c("GO:0048702","embryonic neurocranium morphogenesis", 0.002, 0.142, 6.947, 2.444, 0.0187,0.711,0.433),
c("GO:0048311","mitochondrion distribution", 0.025,-3.646, 1.645, 3.502, 0.0318,0.901,0.436),
c("GO:0046060","dATP metabolic process", 0.001,-3.497,-5.142, 2.207, 0.0134,0.789,0.437),
c("GO:1903071","positive regulation of ER-associated ubiquitin-dependent protein catabolic process", 0.001, 4.542,-2.446, 2.137, 0.0187,0.618,0.440),
c("GO:0033327","Leydig cell differentiation", 0.002,-0.252, 6.634, 2.407, 0.0292,0.686,0.450),
c("GO:0048541","Peyer's patch development", 0.002, 1.607, 6.672, 2.316, 0.0134,0.674,0.470),
c("GO:0060368","regulation of Fc receptor mediated stimulatory signaling pathway", 0.001, 6.210, 1.102, 1.919, 0.0160,0.646,0.480),
c("GO:0006351","transcription, DNA-templated",10.659,-3.806,-3.709, 6.136, 0.0041,0.852,0.480),
c("GO:0072530","purine-containing compound transmembrane transport", 0.016,-4.166, 2.833, 3.304, 0.0213,0.934,0.483),
c("GO:0072531","pyrimidine-containing compound transmembrane transport", 0.019,-4.073, 2.881, 3.391, 0.0422,0.934,0.489),
c("GO:0070255","regulation of mucus secretion", 0.002, 1.155, 4.966, 2.504, 0.0240,0.662,0.494),
c("GO:0038180","nerve growth factor signaling pathway", 0.001, 6.875,-1.582, 2.190, 0.0213,0.729,0.496),
c("GO:0009168","purine ribonucleoside monophosphate biosynthetic process", 1.043,-3.650,-3.225, 5.127, 0.0136,0.734,0.509),
c("GO:0033601","positive regulation of mammary gland epithelial cell proliferation", 0.002, 2.665, 4.905, 2.398, 0.0187,0.604,0.519),
c("GO:0000122","negative regulation of transcription from RNA polymerase II promoter", 0.199, 1.681,-2.816, 4.408, 0.0439,0.725,0.526),
c("GO:0001973","adenosine receptor signaling pathway", 0.005, 7.122,-0.404, 2.792, 0.0292,0.738,0.536),
c("GO:0038063","collagen-activated tyrosine kinase receptor signaling pathway", 0.005, 6.816,-0.631, 2.810, 0.0213,0.742,0.537),
c("GO:0008354","germ cell migration", 0.004,-0.730, 5.666, 2.729, 0.0213,0.700,0.552),
c("GO:0071320","cellular response to cAMP", 0.007, 7.142,-2.642, 2.937, 0.0071,0.810,0.553),
c("GO:0008277","regulation of G-protein coupled receptor protein signaling pathway", 0.024, 6.792, 0.170, 3.494, 0.0209,0.691,0.554),
c("GO:2000178","negative regulation of neural precursor cell proliferation", 0.005, 4.025, 1.444, 2.838, 0.0499,0.775,0.556),
c("GO:0071361","cellular response to ethanol", 0.003, 7.311,-2.794, 2.550, 0.0344,0.820,0.562),
c("GO:0043650","dicarboxylic acid biosynthetic process", 0.530,-5.261,-1.661, 4.833, 0.0344,0.767,0.571),
c("GO:0051412","response to corticosterone", 0.001, 6.485,-3.908, 2.053, 0.0422,0.848,0.572),
c("GO:0034127","regulation of MyD88-independent toll-like receptor signaling pathway", 0.000, 6.487, 1.509, 1.531, 0.0213,0.663,0.590),
c("GO:0002313","mature B cell differentiation involved in immune response", 0.003, 3.280, 4.691, 2.642, 0.0234,0.567,0.592),
c("GO:0032261","purine nucleotide salvage", 0.059,-4.165,-4.110, 3.878, 0.0187,0.768,0.593),
c("GO:0042118","endothelial cell activation", 0.002,-3.033, 0.342, 2.367, 0.0266,0.837,0.600),
c("GO:0003139","secondary heart field specification", 0.002, 0.004, 7.063, 2.358, 0.0240,0.711,0.600),
c("GO:0048387","negative regulation of retinoic acid receptor signaling pathway", 0.002, 6.096, 0.051, 2.394, 0.0213,0.681,0.601),
c("GO:0043249","erythrocyte maturation", 0.002, 2.358, 5.650, 2.461, 0.0292,0.610,0.607),
c("GO:0046130","purine ribonucleoside catabolic process", 0.036,-3.056,-4.878, 3.660, 0.0187,0.745,0.609),
c("GO:0001829","trophectodermal cell differentiation", 0.003,-0.657, 6.646, 2.521, 0.0396,0.689,0.620),
c("GO:0055015","ventricular cardiac muscle cell development", 0.003,-0.775, 6.758, 2.553, 0.0318,0.678,0.622),
c("GO:0002819","regulation of adaptive immune response", 0.025, 6.335, 1.438, 3.513, 0.0184,0.673,0.642),
c("GO:0051446","positive regulation of meiotic cell cycle", 0.007, 3.995, 2.576, 2.925, 0.0422,0.712,0.643),
c("GO:0014807","regulation of somitogenesis", 0.002, 1.564, 6.051, 2.342, 0.0213,0.651,0.660),
c("GO:0035414","negative regulation of catenin import into nucleus", 0.002, 1.162, 1.277, 2.497, 0.0266,0.750,0.669),
c("GO:0018243","protein O-linked glycosylation via threonine", 0.001,-1.054,-6.012, 2.201, 0.0187,0.817,0.676),
c("GO:0046654","tetrahydrofolate biosynthetic process", 0.101,-5.448,-2.197, 4.113, 0.0187,0.765,0.680),
c("GO:0060340","positive regulation of type I interferon-mediated signaling pathway", 0.002, 6.123, 0.385, 2.412, 0.0213,0.608,0.680),
c("GO:0046386","deoxyribose phosphate catabolic process", 0.023,-1.891,-6.345, 3.469, 0.0448,0.843,0.684),
c("GO:2000059","negative regulation of protein ubiquitination involved in ubiquitin-dependent protein catabolic process", 0.003, 2.146,-4.195, 2.603, 0.0318,0.743,0.686),
c("GO:0036066","protein O-linked fucosylation", 0.002,-0.988,-6.193, 2.307, 0.0370,0.815,0.689),
c("GO:0039530","MDA-5 signaling pathway", 0.002, 4.143, 3.197, 2.346, 0.0240,0.527,0.698),
c("GO:0071157","negative regulation of cell cycle arrest", 0.005, 3.360,-0.231, 2.804, 0.0473,0.687,0.699),
c("GO:0035023","regulation of Rho protein signal transduction", 0.125, 6.365, 0.101, 4.206, 0.0400,0.660,0.699));
bjp<-
theme(
panel.border = element_rect(colour = "black", fill = NA, size = 2),
plot.title = element_text(size = 16, face = "bold"),
axis.text.y = element_text(size = 14,face = "bold",color = "black"),
axis.text.x = element_text(size = 14,face = "bold",color = "black"),
axis.title.y = element_text(size = 14,face = "bold"),
axis.title.x = element_text(size = 14,face = "bold"),
legend.text = element_text(size = 14,face = "bold"),
legend.title = element_text(size = 14,face = "bold"),
strip.text.x = element_text(size = 14,face = "bold"),
strip.text.y = element_text(size = 14,face = "bold"),
strip.background = element_rect(colour = "black", size = 2))
one.data <- data.frame(revigo.data);
names(one.data) <- revigo.names;
one.data <- one.data [(one.data$plot_X != "null" & one.data$plot_Y != "null"), ];
one.data$plot_X <- as.numeric( as.character(one.data$plot_X) );
one.data$plot_Y <- as.numeric( as.character(one.data$plot_Y) );
one.data$plot_size <- as.numeric( as.character(one.data$plot_size) );
one.data$log10_p_value <- as.numeric( as.character(one.data$value) );
one.data$frequency <- as.numeric( as.character(one.data$frequency) );
one.data$uniqueness <- as.numeric( as.character(one.data$uniqueness) );
one.data$dispensability <- as.numeric( as.character(one.data$dispensability) );
# --------------------------------------------------------------------------
# This is a table indicating which labels you would like to plot (all data will be plotted, but you need to indicate whether the label should be displayed), make this table manually.
label1 <- c("GO:0070375","ERK5 cascade", 0.001, 3.865,-3.823, 2.041, 0.0134,0.729,0.303)
label2 <- c("GO:0060070","canonical Wnt signaling pathway", 0.061, 4.991,-1.442, 3.893, 0.0066,0.678,0.315)
label3 <- c("GO:0055015","ventricular cardiac muscle cell development", 0.003,-0.775, 6.758, 2.553, 0.0318,0.678,0.622)
label4 <- c("GO:0003139","secondary heart field specification", 0.002, 0.004, 7.063, 2.358, 0.0240,0.711,0.600)
ex <- as.data.frame(rbind(label1, label2, label3), stringsAsFactors = FALSE)
colnames(ex) <- c("term_ID","description","frequency_%","plot_X","plot_Y","plot_size","value","uniqueness","dispensability")
ggplot(data = one.data)+
geom_point( aes( plot_X, plot_Y, fill = log10_p_value, size = plot_size),shape=21,color="black",stroke=0.7, alpha = 0.8) + guides(size=guide_legend(title="No. of genes")) + scale_fill_gradient2(low = muted("red"), mid = "white",
high = muted("blue")) + guides(fill=guide_legend(title="P value"), size=guide_legend(title="No. of genes")) +
#scale_size( range=c(5, 20)) +
labs (y = "Semantic space x", x = "Semantic space y") + theme_bw() + geom_label_repel(ex, mapping = aes(as.numeric(ex$plot_X), as.numeric(ex$plot_Y), label = description),
fontface = 'bold', color = 'black',
box.padding = 0.6, point.padding = 1.6,
size = 3,
# Width of the line segments.
segment.size = 0,
# Strength of the repulsion force.
force = 1)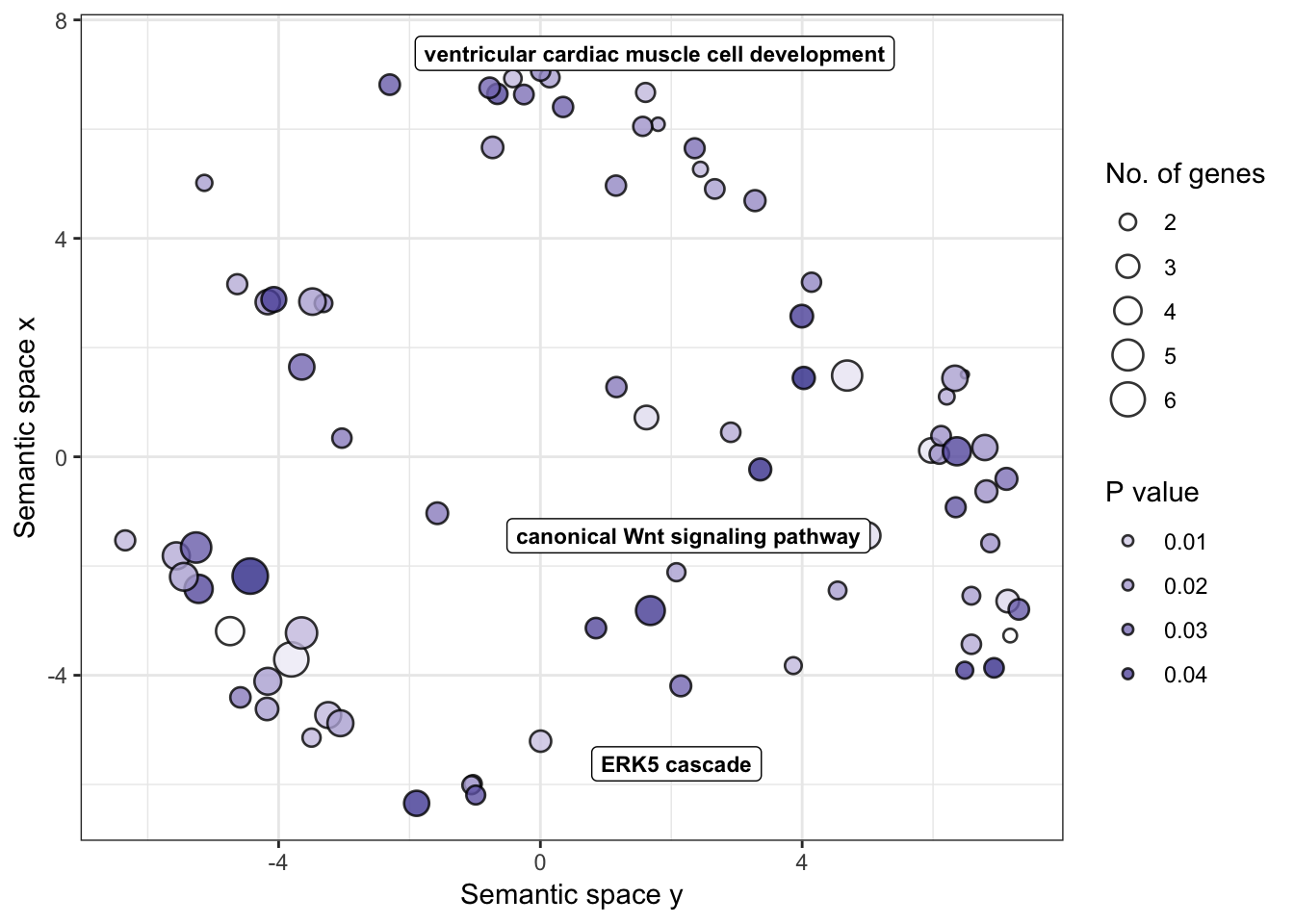
## Revigo liver
revigo.names <- c("term_ID","description","frequency_%","plot_X","plot_Y","plot_size","value","uniqueness","dispensability");
revigo.data <- rbind(c("GO:0002576","platelet degranulation", 0.003, 0.496,-3.539, 2.519, 0.0000,0.842,0.000),
c("GO:0008152","metabolic process",75.387, 0.813,-0.564, 6.986, 0.0308,0.998,0.000),
c("GO:0009820","alkaloid metabolic process", 0.006, 4.043, 1.159, 2.869, 0.0000,0.947,0.000),
c("GO:0042632","cholesterol homeostasis", 0.019,-3.975, 2.639, 3.381, 0.0000,0.818,0.000),
c("GO:0019835","cytolysis", 0.044, 0.583,-0.452, 3.749, 0.0007,0.971,0.018),
c("GO:0001887","selenium compound metabolic process", 0.008, 0.543,-0.357, 2.990, 0.0189,0.963,0.019),
c("GO:0042157","lipoprotein metabolic process", 0.210, 0.683,-0.584, 4.431, 0.0116,0.957,0.028),
c("GO:0017144","drug metabolic process", 0.058, 0.564,-0.386, 3.868, 0.0392,0.959,0.035),
c("GO:0070989","oxidative demethylation", 0.011, 1.605, 0.368, 3.133, 0.0064,0.912,0.041),
c("GO:0019740","nitrogen utilization", 0.085, 0.842,-0.775, 4.038, 0.0189,0.932,0.050),
c("GO:0019695","choline metabolic process", 0.016, 0.978, 0.110, 3.301, 0.0060,0.962,0.063),
c("GO:0048625","myoblast fate commitment", 0.001, 0.704,-6.082, 2.100, 0.0189,0.822,0.085),
c("GO:0006368","transcription elongation from RNA polymerase II promoter", 0.082, 1.989, 1.876, 4.021, 0.0139,0.890,0.085),
c("GO:0051081","nuclear envelope disassembly", 0.002, 0.688,-0.443, 2.338, 0.0116,0.883,0.087),
c("GO:0006957","complement activation, alternative pathway", 0.000,-6.310, 2.907, 1.771, 0.0003,0.689,0.121),
c("GO:0017187","peptidyl-glutamic acid carboxylation", 0.006,-0.167, 1.374, 2.865, 0.0011,0.918,0.125),
c("GO:0006629","lipid metabolic process", 3.522, 3.786, 1.634, 5.655, 0.0144,0.876,0.126),
c("GO:0034384","high-density lipoprotein particle clearance", 0.002,-3.947,-4.331, 2.294, 0.0001,0.733,0.128),
c("GO:1904683","regulation of metalloendopeptidase activity", 0.001,-3.073, 3.701, 2.210, 0.0017,0.790,0.131),
c("GO:0006641","triglyceride metabolic process", 0.038, 3.327, 1.871, 3.687, 0.0001,0.832,0.140),
c("GO:0051647","nucleus localization", 0.012,-0.140,-3.521, 3.198, 0.0117,0.927,0.141),
c("GO:0042738","exogenous drug catabolic process", 0.001,-1.097, 3.996, 2.193, 0.0009,0.834,0.156),
c("GO:0070141","response to UV-A", 0.002,-3.392, 1.485, 2.369, 0.0017,0.918,0.166),
c("GO:0006768","biotin metabolic process", 0.081, 5.718, 3.455, 4.018, 0.0007,0.805,0.167),
c("GO:1901160","primary amino compound metabolic process", 0.015, 4.703, 1.279, 3.290, 0.0455,0.944,0.178),
c("GO:0043627","response to estrogen", 0.010,-3.637, 1.577, 3.128, 0.0379,0.912,0.184),
c("GO:0099133","ATP hydrolysis coupled anion transmembrane transport", 0.271,-0.029,-4.342, 4.541, 0.0152,0.919,0.184),
c("GO:0010646","regulation of cell communication", 0.929,-6.450, 3.995, 5.076, 0.0173,0.784,0.190),
c("GO:0007041","lysosomal transport", 0.017, 0.052,-3.377, 3.341, 0.0395,0.947,0.196),
c("GO:0042158","lipoprotein biosynthetic process", 0.192, 0.957, 0.864, 4.391, 0.0189,0.919,0.199),
c("GO:1990535","neuron projection maintenance", 0.000, 0.267,-0.657, 1.301, 0.0275,0.897,0.200),
c("GO:0052652","cyclic purine nucleotide metabolic process", 0.094, 4.152, 4.182, 4.082, 0.0457,0.804,0.234),
c("GO:0006390","transcription from mitochondrial promoter", 0.005, 0.802, 1.042, 2.794, 0.0484,0.915,0.235),
c("GO:0006600","creatine metabolic process", 0.002, 5.427, 2.495, 2.328, 0.0374,0.857,0.244),
c("GO:0042127","regulation of cell proliferation", 0.313,-5.699, 3.430, 4.603, 0.0229,0.785,0.259),
c("GO:0016540","protein autoprocessing", 0.011,-0.432, 1.323, 3.147, 0.0191,0.924,0.274),
c("GO:0006476","protein deacetylation", 0.072,-0.361, 1.247, 3.967, 0.0455,0.913,0.278),
c("GO:0055114","oxidation-reduction process",15.060, 4.858, 2.263, 6.286, 0.0206,0.872,0.300),
c("GO:0015886","heme transport", 0.066,-0.143,-4.692, 3.927, 0.0044,0.860,0.302),
c("GO:0006461","protein complex assembly", 0.960, 0.086,-0.705, 5.091, 0.0285,0.920,0.312),
c("GO:0060052","neurofilament cytoskeleton organization", 0.002, 1.098,-0.364, 2.501, 0.0374,0.883,0.319),
c("GO:0010918","positive regulation of mitochondrial membrane potential", 0.001,-2.305, 1.655, 2.164, 0.0044,0.851,0.327),
c("GO:0033572","transferrin transport", 0.002,-0.385,-3.961, 2.449, 0.0180,0.915,0.333),
c("GO:0006524","alanine catabolic process", 0.025, 5.584, 3.621, 3.509, 0.0009,0.800,0.338),
c("GO:0097267","omega-hydroxylase P450 pathway", 0.000, 4.727, 2.654, 1.000, 0.0017,0.851,0.339),
c("GO:0019627","urea metabolic process", 0.051, 4.765, 2.756, 3.820, 0.0455,0.846,0.339),
c("GO:1903830","magnesium ion transmembrane transport", 0.088, 0.021,-4.086, 4.053, 0.0339,0.938,0.340),
c("GO:1904179","positive regulation of adipose tissue development", 0.001,-5.562,-3.251, 1.996, 0.0009,0.657,0.344),
c("GO:0007584","response to nutrient", 0.026,-3.941, 1.599, 3.530, 0.0010,0.865,0.345),
c("GO:0090400","stress-induced premature senescence", 0.002,-4.185,-2.548, 2.350, 0.0484,0.752,0.346),
c("GO:0042908","xenobiotic transport", 0.028,-0.145,-4.010, 3.556, 0.0064,0.876,0.348),
c("GO:0060021","palate development", 0.018,-0.008,-5.732, 3.363, 0.0445,0.883,0.355),
c("GO:0033860","regulation of NAD(P)H oxidase activity", 0.001,-3.702, 4.403, 2.057, 0.0275,0.831,0.356),
c("GO:0045725","positive regulation of glycogen biosynthetic process", 0.003,-0.905, 5.340, 2.547, 0.0039,0.686,0.357),
c("GO:0001907","killing by symbiont of host cells", 0.008,-3.276, 2.257, 3.001, 0.0009,0.818,0.362),
c("GO:0016310","phosphorylation", 7.764, 2.492, 2.058, 5.998, 0.0064,0.912,0.371),
c("GO:0050892","intestinal absorption", 0.006,-0.941,-7.157, 2.913, 0.0016,0.782,0.379),
c("GO:1900107","regulation of nodal signaling pathway", 0.002,-6.525, 3.443, 2.316, 0.0275,0.740,0.380),
c("GO:0008090","retrograde axonal transport", 0.003, 0.413,-4.290, 2.521, 0.0374,0.827,0.390),
c("GO:0008203","cholesterol metabolic process", 0.028, 5.004, 2.844, 3.554, 0.0004,0.835,0.402),
c("GO:0070301","cellular response to hydrogen peroxide", 0.019,-5.078, 2.109, 3.397, 0.0023,0.823,0.402),
c("GO:0007628","adult walking behavior", 0.008,-0.864,-5.875, 3.000, 0.0288,0.814,0.411),
c("GO:0043433","negative regulation of sequence-specific DNA binding transcription factor activity", 0.031,-3.638, 4.535, 3.600, 0.0023,0.715,0.412),
c("GO:0051340","regulation of ligase activity", 0.002,-2.775, 3.103, 2.391, 0.0455,0.849,0.421),
c("GO:0097190","apoptotic signaling pathway", 0.117,-4.388, 4.477, 4.177, 0.0357,0.702,0.427),
c("GO:0006869","lipid transport", 0.270,-0.359,-4.931, 4.539, 0.0084,0.825,0.430),
c("GO:0045717","negative regulation of fatty acid biosynthetic process", 0.007, 0.731, 5.462, 2.935, 0.0030,0.667,0.434),
c("GO:0051004","regulation of lipoprotein lipase activity", 0.003,-2.788, 2.773, 2.587, 0.0089,0.840,0.435),
c("GO:0006536","glutamate metabolic process", 0.201, 5.539, 3.254, 4.412, 0.0020,0.816,0.441),
c("GO:0018279","protein N-linked glycosylation via asparagine", 0.015, 1.838, 3.560, 3.284, 0.0410,0.833,0.446),
c("GO:0046185","aldehyde catabolic process", 0.048, 4.214, 2.777, 3.790, 0.0064,0.832,0.451),
c("GO:0032071","regulation of endodeoxyribonuclease activity", 0.003,-3.421, 4.017, 2.537, 0.0374,0.791,0.451),
c("GO:0008202","steroid metabolic process", 0.161, 3.336, 1.762, 4.315, 0.0005,0.851,0.451),
c("GO:0071156","regulation of cell cycle arrest", 0.013,-2.547, 3.556, 3.230, 0.0275,0.745,0.452),
c("GO:0071372","cellular response to follicle-stimulating hormone stimulus", 0.001,-4.403, 1.848, 2.220, 0.0118,0.859,0.460),
c("GO:0042940","D-amino acid transport", 0.003,-0.063,-4.288, 2.562, 0.0275,0.873,0.471),
c("GO:1903427","negative regulation of reactive oxygen species biosynthetic process", 0.002,-3.996, 3.252, 2.486, 0.0044,0.777,0.477),
c("GO:0042493","response to drug", 0.266,-5.108, 2.172, 4.534, 0.0449,0.863,0.477),
c("GO:0001889","liver development", 0.023,-1.101,-6.479, 3.471, 0.0009,0.766,0.478),
c("GO:0006545","glycine biosynthetic process", 0.079, 5.573, 3.375, 4.005, 0.0275,0.814,0.479),
c("GO:0019216","regulation of lipid metabolic process", 0.095,-2.218, 5.684, 4.086, 0.0052,0.745,0.482),
c("GO:0006591","ornithine metabolic process", 0.088, 5.478, 3.118, 4.052, 0.0484,0.825,0.483),
c("GO:0046838","phosphorylated carbohydrate dephosphorylation", 0.064, 1.908, 2.032, 3.915, 0.0455,0.926,0.484),
c("GO:0006739","NADP metabolic process", 0.357, 4.701, 4.332, 4.661, 0.0063,0.793,0.485),
c("GO:0006520","cellular amino acid metabolic process", 5.591, 5.069, 3.403, 5.856, 0.0148,0.779,0.487),
c("GO:2000343","positive regulation of chemokine (C-X-C motif) ligand 2 production", 0.001,-5.534,-2.610, 1.959, 0.0275,0.680,0.489),
c("GO:0021814","cell motility involved in cerebral cortex radial glia guided migration", 0.001,-0.848,-6.605, 2.238, 0.0189,0.729,0.492),
c("GO:0022900","electron transport chain", 0.564, 3.324, 2.185, 4.860, 0.0186,0.848,0.493),
c("GO:0090277","positive regulation of peptide hormone secretion", 0.013,-4.678, 0.230, 3.226, 0.0004,0.601,0.496),
c("GO:0015942","formate metabolic process", 0.065, 5.321, 3.235, 3.921, 0.0017,0.832,0.499),
c("GO:0046329","negative regulation of JNK cascade", 0.007,-4.599, 5.445, 2.940, 0.0063,0.605,0.500),
c("GO:0001503","ossification", 0.074,-0.710,-6.573, 3.979, 0.0413,0.791,0.504),
c("GO:0006898","receptor-mediated endocytosis", 0.095, 0.278,-4.169, 4.086, 0.0004,0.935,0.511),
c("GO:0090239","regulation of histone H4 acetylation", 0.003,-3.175, 3.088, 2.531, 0.0455,0.769,0.511),
c("GO:0060059","embryonic retina morphogenesis in camera-type eye", 0.003,-0.909,-6.515, 2.573, 0.0029,0.790,0.512),
c("GO:1902459","positive regulation of stem cell population maintenance", 0.001,-4.906,-2.513, 2.236, 0.0275,0.663,0.517),
c("GO:1902959","regulation of aspartic-type endopeptidase activity involved in amyloid precursor protein catabolic process", 0.000,-2.854, 3.852, 1.756, 0.0455,0.802,0.517),
c("GO:0031100","animal organ regeneration", 0.005,-1.364,-6.064, 2.792, 0.0402,0.788,0.526),
c("GO:0061620","glycolytic process through glucose-6-phosphate", 0.002, 5.004, 4.281, 2.290, 0.0455,0.798,0.529),
c("GO:0002679","respiratory burst involved in defense response", 0.001,-3.703, 1.450, 2.258, 0.0374,0.816,0.535),
c("GO:0034504","protein localization to nucleus", 0.129,-0.313,-4.647, 4.218, 0.0210,0.899,0.539),
c("GO:0014911","positive regulation of smooth muscle cell migration", 0.003,-5.348,-0.209, 2.620, 0.0461,0.695,0.540),
c("GO:0061138","morphogenesis of a branching epithelium", 0.042,-1.117,-6.445, 3.730, 0.0021,0.775,0.541),
c("GO:0010793","regulation of mRNA export from nucleus", 0.003,-3.669,-1.160, 2.625, 0.0275,0.749,0.541),
c("GO:0072049","comma-shaped body morphogenesis", 0.001,-1.363,-5.960, 2.173, 0.0189,0.801,0.543),
c("GO:0046498","S-adenosylhomocysteine metabolic process", 0.066, 5.725, 3.823, 3.925, 0.0275,0.808,0.544),
c("GO:0045620","negative regulation of lymphocyte differentiation", 0.008,-3.807,-2.279, 3.018, 0.0029,0.590,0.549),
c("GO:0019626","short-chain fatty acid catabolic process", 0.024, 5.002, 3.731, 3.490, 0.0189,0.779,0.556),
c("GO:0034356","NAD biosynthesis via nicotinamide riboside salvage pathway", 0.004, 4.983, 4.281, 2.691, 0.0235,0.828,0.560),
c("GO:0046620","regulation of organ growth", 0.020,-4.335,-3.406, 3.403, 0.0044,0.688,0.561),
c("GO:0006778","porphyrin-containing compound metabolic process", 0.457, 5.545, 2.444, 4.768, 0.0453,0.896,0.561),
c("GO:0010269","response to selenium ion", 0.001,-3.417, 1.446, 1.991, 0.0189,0.892,0.564),
c("GO:0090335","regulation of brown fat cell differentiation", 0.003,-3.134,-2.585, 2.531, 0.0274,0.735,0.566),
c("GO:0000098","sulfur amino acid catabolic process", 0.008, 5.398, 3.462, 3.030, 0.0064,0.815,0.566),
c("GO:1905668","positive regulation of protein localization to endosome", 0.001,-4.713,-0.861, 2.079, 0.0275,0.732,0.569),
c("GO:0031016","pancreas development", 0.021,-1.275,-6.229, 3.438, 0.0453,0.774,0.573),
c("GO:0071397","cellular response to cholesterol", 0.001,-4.311, 1.853, 2.272, 0.0191,0.857,0.574),
c("GO:0060712","spongiotrophoblast layer development", 0.003,-1.057,-6.380, 2.533, 0.0191,0.790,0.578),
c("GO:0031659","positive regulation of cyclin-dependent protein serine/threonine kinase activity involved in G1/S transition of mitotic cell cycle", 0.002,-3.538, 4.926, 2.350, 0.0275,0.670,0.578),
c("GO:1903896","positive regulation of IRE1-mediated unfolded protein response", 0.001,-6.708, 3.143, 1.857, 0.0275,0.671,0.580),
c("GO:0071560","cellular response to transforming growth factor beta stimulus", 0.050,-4.466, 1.856, 3.804, 0.0127,0.832,0.582),
c("GO:0035733","hepatic stellate cell activation", 0.000, 0.492,-0.383, 1.748, 0.0455,0.898,0.584),
c("GO:0034763","negative regulation of transmembrane transport", 0.014,-4.429,-0.887, 3.251, 0.0274,0.743,0.585),
c("GO:0042167","heme catabolic process", 0.002, 5.014, 2.712, 2.408, 0.0374,0.834,0.586),
c("GO:0006853","carnitine shuttle", 0.000,-0.241,-4.610, 1.114, 0.0089,0.860,0.587),
c("GO:0007411","axon guidance", 0.066,-3.249,-3.116, 3.930, 0.0223,0.617,0.587),
c("GO:0048013","ephrin receptor signaling pathway", 0.016,-6.783, 3.560, 3.308, 0.0309,0.752,0.593),
c("GO:0034340","response to type I interferon", 0.008,-5.227, 1.865, 3.007, 0.0117,0.821,0.598),
c("GO:0031325","positive regulation of cellular metabolic process", 1.004,-6.818, 4.034, 5.110, 0.0233,0.668,0.602),
c("GO:0042177","negative regulation of protein catabolic process", 0.025,-2.311, 5.307, 3.513, 0.0177,0.698,0.606),
c("GO:1900016","negative regulation of cytokine production involved in inflammatory response", 0.002,-5.942,-1.181, 2.391, 0.0191,0.603,0.606),
c("GO:2000463","positive regulation of excitatory postsynaptic potential", 0.003,-6.002, 3.548, 2.606, 0.0220,0.638,0.607),
c("GO:0098902","regulation of membrane depolarization during action potential", 0.001,-3.098, 2.354, 2.188, 0.0455,0.827,0.608),
c("GO:1902237","positive regulation of endoplasmic reticulum stress-induced intrinsic apoptotic signaling pathway", 0.002,-6.892, 3.889, 2.324, 0.0089,0.647,0.617),
c("GO:0008635","activation of cysteine-type endopeptidase activity involved in apoptotic process by cytochrome c", 0.001,-5.340, 4.602, 2.279, 0.0484,0.665,0.619),
c("GO:2000271","positive regulation of fibroblast apoptotic process", 0.002,-6.128, 4.110, 2.320, 0.0374,0.726,0.621),
c("GO:0050730","regulation of peptidyl-tyrosine phosphorylation", 0.045,-2.927, 5.206, 3.759, 0.0115,0.746,0.627),
c("GO:0014068","positive regulation of phosphatidylinositol 3-kinase signaling", 0.013,-6.835, 3.378, 3.216, 0.0477,0.656,0.627),
c("GO:0032148","activation of protein kinase B activity", 0.004,-3.889, 4.818, 2.738, 0.0411,0.689,0.628),
c("GO:0000122","negative regulation of transcription from RNA polymerase II promoter", 0.199,-3.286, 4.570, 4.408, 0.0343,0.704,0.628),
c("GO:2000659","regulation of interleukin-1-mediated signaling pathway", 0.001,-6.610, 3.398, 2.117, 0.0275,0.727,0.629),
c("GO:0043392","negative regulation of DNA binding", 0.014,-3.451, 2.990, 3.256, 0.0324,0.830,0.631),
c("GO:0042426","choline catabolic process", 0.001, 4.091, 2.217, 2.230, 0.0275,0.898,0.632),
c("GO:1900119","positive regulation of execution phase of apoptosis", 0.003,-5.254, 3.939, 2.528, 0.0152,0.720,0.635),
c("GO:0046487","glyoxylate metabolic process", 0.049, 5.512, 3.224, 3.794, 0.0484,0.832,0.635),
c("GO:0030917","midbrain-hindbrain boundary development", 0.003,-0.916,-6.603, 2.576, 0.0064,0.776,0.636),
c("GO:0031663","lipopolysaccharide-mediated signaling pathway", 0.010,-6.114, 3.001, 3.128, 0.0363,0.713,0.638),
c("GO:1901679","nucleotide transmembrane transport", 0.011,-0.193,-4.465, 3.131, 0.0374,0.862,0.638),
c("GO:0032933","SREBP signaling pathway", 0.008,-6.008, 4.192, 2.989, 0.0484,0.656,0.640),
c("GO:2000646","positive regulation of receptor catabolic process", 0.001,-2.717, 5.596, 1.881, 0.0189,0.731,0.642),
c("GO:0071276","cellular response to cadmium ion", 0.009,-4.925, 2.123, 3.077, 0.0040,0.848,0.644),
c("GO:0000060","protein import into nucleus, translocation", 0.012,-0.358,-4.229, 3.175, 0.0286,0.853,0.645),
c("GO:0006548","histidine catabolic process", 0.052, 5.484, 3.853, 3.828, 0.0044,0.789,0.649),
c("GO:0034638","phosphatidylcholine catabolic process", 0.002, 4.403, 3.693, 2.328, 0.0374,0.793,0.652),
c("GO:1901523","icosanoid catabolic process", 0.000, 5.088, 3.154, 1.732, 0.0189,0.841,0.653),
c("GO:0006559","L-phenylalanine catabolic process", 0.031, 5.469, 3.690, 3.596, 0.0118,0.794,0.657),
c("GO:0051642","centrosome localization", 0.007,-0.128,-3.483, 2.973, 0.0190,0.928,0.658),
c("GO:0010040","response to iron(II) ion", 0.002,-4.034, 1.687, 2.407, 0.0189,0.886,0.658),
c("GO:0000042","protein targeting to Golgi", 0.021,-0.296,-4.424, 3.422, 0.0374,0.900,0.670),
c("GO:0090201","negative regulation of release of cytochrome c from mitochondria", 0.003,-5.536, 3.408, 2.591, 0.0285,0.651,0.672),
c("GO:0006855","drug transmembrane transport", 0.189,-4.897,-2.034, 4.384, 0.0138,0.752,0.672),
c("GO:0009887","animal organ morphogenesis", 0.264,-1.342,-6.260, 4.530, 0.0333,0.742,0.673),
c("GO:0006879","cellular iron ion homeostasis", 0.110,-1.935, 4.780, 4.148, 0.0002,0.750,0.674),
c("GO:0030178","negative regulation of Wnt signaling pathway", 0.037,-5.011, 4.290, 3.681, 0.0367,0.636,0.676),
c("GO:0007262","STAT protein import into nucleus", 0.001,-5.800,-0.845, 2.223, 0.0484,0.729,0.677),
c("GO:0045739","positive regulation of DNA repair", 0.013,-5.991, 3.701, 3.205, 0.0123,0.635,0.678),
c("GO:0034142","toll-like receptor 4 signaling pathway", 0.006,-6.777, 3.183, 2.892, 0.0191,0.642,0.679),
c("GO:0046326","positive regulation of glucose import", 0.007,-6.120,-0.943, 2.979, 0.0287,0.694,0.680),
c("GO:0042773","ATP synthesis coupled electron transport", 0.221, 4.104, 4.074, 4.452, 0.0456,0.794,0.685),
c("GO:0010886","positive regulation of cholesterol storage", 0.002,-5.084,-0.414, 2.320, 0.0374,0.711,0.685),
c("GO:0001568","blood vessel development", 0.136,-1.287,-6.290, 4.243, 0.0452,0.758,0.689),
c("GO:0006505","GPI anchor metabolic process", 0.102, 3.442, 3.385, 4.115, 0.0456,0.805,0.690),
c("GO:0046439","L-cysteine metabolic process", 0.039, 6.129, 3.195, 3.699, 0.0374,0.827,0.692),
c("GO:0051897","positive regulation of protein kinase B signaling", 0.016,-6.848, 3.394, 3.304, 0.0485,0.653,0.692),
c("GO:0032287","peripheral nervous system myelin maintenance", 0.002,-0.055,-6.738, 2.468, 0.0374,0.750,0.693),
c("GO:1904468","negative regulation of tumor necrosis factor secretion", 0.001,-3.544,-4.697, 1.982, 0.0374,0.625,0.695),
c("GO:1903874","ferrous iron transmembrane transport", 0.028, 0.089,-4.022, 3.563, 0.0374,0.931,0.700));
one.data <- data.frame(revigo.data);
names(one.data) <- revigo.names;
one.data <- one.data [(one.data$plot_X != "null" & one.data$plot_Y != "null"), ];
one.data$plot_X <- as.numeric( as.character(one.data$plot_X) );
one.data$plot_Y <- as.numeric( as.character(one.data$plot_Y) );
one.data$plot_size <- as.numeric( as.character(one.data$plot_size) );
one.data$log10_p_value <- as.numeric( as.character(one.data$value) );
one.data$frequency <- as.numeric( as.character(one.data$frequency) );
one.data$uniqueness <- as.numeric( as.character(one.data$uniqueness) );
one.data$dispensability <- as.numeric( as.character(one.data$dispensability) );
# --------------------------------------------------------------------------
# This is a table indicating which labels you would like to plot (all data will be plotted, but you need to indicate whether the label should be displayed), make this table manually.
label1 <- c("GO:0042632","cholesterol homeostasis", 0.019,-3.975, 2.639, 3.381, 0.0000,0.818,0.000)
label2 <- c("GO:0017144","drug metabolic process", 0.058, 0.564,-0.386, 3.868, 0.0392,0.959,0.035)
label3 <- c("GO:0006629","lipid metabolic process", 3.522, 3.786, 1.634, 5.655, 0.0144,0.876,0.126)
ex <- as.data.frame(rbind(label1, label2, label3), stringsAsFactors = FALSE)
colnames(ex) <- c("term_ID","description","frequency_%","plot_X","plot_Y","plot_size","value","uniqueness","dispensability")
ggplot(data = one.data)+
geom_point( aes( plot_X, plot_Y, fill = log10_p_value, size = plot_size),shape=21,color="black",stroke=0.7, alpha = 0.8) + guides(size=guide_legend(title="No. of genes")) + scale_fill_gradient2(low = muted("red"), mid = "white",
high = muted("blue")) + guides(fill=guide_legend(title="P value"), size=guide_legend(title="No. of genes")) +
#scale_size( range=c(5, 20)) +
labs (y = "Semantic space x", x = "Semantic space y") + theme_bw() + geom_label_repel(ex, mapping = aes(as.numeric(ex$plot_X), as.numeric(ex$plot_Y), label = description),
fontface = 'bold', color = 'black',
box.padding = 0.6, point.padding = 1.6,
size = 3,
# Width of the line segments.
segment.size = 0,
# Strength of the repulsion force.
force = 1)