Weight_change_preliminary_analysis
Lauren Blake
2018-09-17
Last updated: 2018-11-15
workflowr checks: (Click a bullet for more information)-
✔ R Markdown file: up-to-date
Great! Since the R Markdown file has been committed to the Git repository, you know the exact version of the code that produced these results.
-
✔ Environment: empty
Great job! The global environment was empty. Objects defined in the global environment can affect the analysis in your R Markdown file in unknown ways. For reproduciblity it’s best to always run the code in an empty environment.
-
✔ Seed:
set.seed(12345)The command
set.seed(12345)was run prior to running the code in the R Markdown file. Setting a seed ensures that any results that rely on randomness, e.g. subsampling or permutations, are reproducible. -
✔ Session information: recorded
Great job! Recording the operating system, R version, and package versions is critical for reproducibility.
-
Great! You are using Git for version control. Tracking code development and connecting the code version to the results is critical for reproducibility. The version displayed above was the version of the Git repository at the time these results were generated.✔ Repository version: 7bcd123
Note that you need to be careful to ensure that all relevant files for the analysis have been committed to Git prior to generating the results (you can usewflow_publishorwflow_git_commit). workflowr only checks the R Markdown file, but you know if there are other scripts or data files that it depends on. Below is the status of the Git repository when the results were generated:
Note that any generated files, e.g. HTML, png, CSS, etc., are not included in this status report because it is ok for generated content to have uncommitted changes.Ignored files: Ignored: .DS_Store Ignored: analysis/.DS_Store Ignored: analysis/VennDiagram2018-07-24_06-55-46.log Ignored: analysis/VennDiagram2018-07-24_06-56-13.log Ignored: analysis/VennDiagram2018-07-24_06-56-50.log Ignored: analysis/VennDiagram2018-07-24_06-58-41.log Ignored: analysis/VennDiagram2018-07-24_07-00-07.log Ignored: analysis/VennDiagram2018-07-24_07-00-42.log Ignored: analysis/VennDiagram2018-07-24_07-01-08.log Ignored: analysis/VennDiagram2018-08-17_15-13-24.log Ignored: analysis/VennDiagram2018-08-17_15-13-30.log Ignored: analysis/VennDiagram2018-08-17_15-15-06.log Ignored: analysis/VennDiagram2018-08-17_15-16-01.log Ignored: analysis/VennDiagram2018-08-17_15-17-51.log Ignored: analysis/VennDiagram2018-08-17_15-18-42.log Ignored: analysis/VennDiagram2018-08-17_15-19-21.log Ignored: analysis/VennDiagram2018-08-20_09-07-57.log Ignored: analysis/VennDiagram2018-08-20_09-08-37.log Ignored: analysis/VennDiagram2018-08-26_19-54-03.log Ignored: analysis/VennDiagram2018-08-26_20-47-08.log Ignored: analysis/VennDiagram2018-08-26_20-49-49.log Ignored: analysis/VennDiagram2018-08-27_00-04-36.log Ignored: analysis/VennDiagram2018-08-27_00-09-27.log Ignored: analysis/VennDiagram2018-08-27_00-13-57.log Ignored: analysis/VennDiagram2018-08-27_00-16-32.log Ignored: analysis/VennDiagram2018-08-27_10-00-25.log Ignored: analysis/VennDiagram2018-08-28_06-03-13.log Ignored: analysis/VennDiagram2018-08-28_06-03-14.log Ignored: analysis/VennDiagram2018-08-28_06-05-50.log Ignored: analysis/VennDiagram2018-08-28_06-06-58.log Ignored: analysis/VennDiagram2018-08-28_06-10-12.log Ignored: analysis/VennDiagram2018-08-28_06-10-13.log Ignored: analysis/VennDiagram2018-08-28_06-18-29.log Ignored: analysis/VennDiagram2018-08-28_07-22-26.log Ignored: analysis/VennDiagram2018-08-28_07-22-27.log Ignored: analysis/VennDiagram2018-08-28_13-05-27.log Ignored: analysis/VennDiagram2018-09-12_01-45-59.log Ignored: analysis/VennDiagram2018-09-12_01-49-31.log Ignored: analysis/VennDiagram2018-09-12_01-58-11.log Ignored: analysis/VennDiagram2018-09-12_01-59-46.log Ignored: analysis/VennDiagram2018-09-12_02-08-07.log Ignored: analysis/VennDiagram2018-09-12_02-08-56.log Ignored: analysis/VennDiagram2018-11-15_14-20-08.log Ignored: analysis/VennDiagram2018-11-15_14-20-15.log Ignored: analysis/VennDiagram2018-11-15_14-20-23.log Ignored: analysis/VennDiagram2018-11-15_14-21-14.log Ignored: analysis/VennDiagram2018-11-15_14-21-57.log Ignored: analysis/VennDiagram2018-11-15_14-33-34.log Ignored: analysis/VennDiagram2018-11-15_14-36-19.log Ignored: analysis/VennDiagram2018-11-15_14-48-41.log Ignored: analysis/VennDiagram2018-11-15_14-48-42.log Ignored: analysis/VennDiagram2018-11-15_15-03-35.log Ignored: analysis/VennDiagram2018-11-15_15-03-55.log Ignored: analysis/VennDiagram2018-11-15_15-07-05.log Ignored: analysis/VennDiagram2018-11-15_15-07-25.log Ignored: analysis/VennDiagram2018-11-15_15-09-29.log Ignored: analysis/VennDiagram2018-11-15_15-09-48.log Ignored: analysis/VennDiagram2018-11-15_15-14-30.log Ignored: analysis/VennDiagram2018-11-15_15-15-25.log Ignored: analysis/VennDiagram2018-11-15_15-16-04.log Ignored: data/DAVID_2covar/ Ignored: data/DAVID_results/ Ignored: data/Eigengenes/ Ignored: data/aux_info/ Ignored: data/hg_38/ Ignored: data/libParams/ Ignored: data/logs/ Ignored: docs/VennDiagram2018-07-24_06-55-46.log Ignored: docs/VennDiagram2018-07-24_06-56-13.log Ignored: docs/VennDiagram2018-07-24_06-56-50.log Ignored: docs/VennDiagram2018-07-24_06-58-41.log Ignored: docs/VennDiagram2018-07-24_07-00-07.log Ignored: docs/VennDiagram2018-07-24_07-00-42.log Ignored: docs/VennDiagram2018-07-24_07-01-08.log Ignored: docs/figure/.DS_Store Ignored: output/.DS_Store Untracked files: Untracked: docs/figure/time_two_covar.Rmd/ Unstaged changes: Modified: analysis/time_two_covar.Rmd
Expand here to see past versions:
| File | Version | Author | Date | Message |
|---|---|---|---|---|
| Rmd | 7bcd123 | Lauren Blake | 2018-11-15 | Nov 15 weight info |
Introduction
The goal of this script is to identify the individuals that weight relapsed and the amount of change they underwent.
Get weight change data
# Load libraries
library(ggplot2)Warning: package 'ggplot2' was built under R version 3.4.4library(cowplot)Warning: package 'cowplot' was built under R version 3.4.4
Attaching package: 'cowplot'The following object is masked from 'package:ggplot2':
ggsave# Get weight change data
weight_change <- read.csv("../data/weight_change.csv")Summary of weight change data
# Subset to time point 3
weight_change_t3 <- weight_change[weight_change$time == 3,]
summary(weight_change_t3) BAN_ID time Weight Weight_change
Min. :2201 Min. :3 Min. : 71.5 Min. :-14.500
1st Qu.:2218 1st Qu.:3 1st Qu.:102.8 1st Qu.: 0.900
Median :2239 Median :3 Median :108.0 Median : 7.100
Mean :2237 Mean :3 Mean :110.0 Mean : 7.601
3rd Qu.:2256 3rd Qu.:3 3rd Qu.:118.4 3rd Qu.: 10.800
Max. :2274 Max. :3 Max. :148.9 Max. : 45.200
NA's :14 NA's :14
Relapse_t3 Relapse_t4 Relapse_t5
Min. :0.0000 Min. :0 Min. :0.0
1st Qu.:0.0000 1st Qu.:0 1st Qu.:0.0
Median :0.0000 Median :0 Median :0.5
Mean :0.1951 Mean :0 Mean :0.5
3rd Qu.:0.0000 3rd Qu.:0 3rd Qu.:1.0
Max. :1.0000 Max. :0 Max. :1.0
NA's :14 NA's :49 NA's :49 # How many are negative and how many are neutral/positive weight gain?
summary(weight_change_t3$Weight_change <0) Mode FALSE TRUE NA's
logical 33 8 14 weight_change_t3[,5] <- weight_change_t3$Weight_change <0
colnames(weight_change_t3) <- c("BAN_ID", "time", "Weight", "Weight_change", "weight_relapse")
# Weight change from T2 to T3
weight_change_t3 <- weight_change_t3[complete.cases(weight_change_t3), ]
weight_change_t3_plot <- ggplot(weight_change_t3, aes(weight_relapse, Weight_change)) + geom_boxplot() + xlab("Lost weight from T2 to T3") + ylab("Weight change from T2 to T3")
save_plot("/Users/laurenblake/Dropbox/Figures/weight_change_t3.png", weight_change_t3_plot, base_aspect_ratio = 1)Weights over time, stratified by weight loss at T3
# Subset to time point 1
weight_change_t1 <- weight_change[weight_change$time == 1, 1:5]
# How many are negative and how many are neutral/positive weight gain?
summary(weight_change_t1$Relapse_t3 >0) Mode FALSE TRUE NA's
logical 33 8 14 weight_change_t1[,6] <- weight_change_t1$Relapse_t3 >0
colnames(weight_change_t1) <- c("BAN_ID", "time", "Weight", "Weight_change", "Relapse_t3", "Relapse_notes")
weight_change_t1 <- weight_change_t1[complete.cases(weight_change_t1), ]
# Plot weight at T1
weight_t1_plot <- ggplot(weight_change_t1, aes(Relapse_notes, Weight)) + geom_boxplot() + xlab("Lost weight from T2 to T3") + ylab("Weight") + ggtitle("Weight at T1")
save_plot("/Users/laurenblake/Dropbox/Figures/weight_t1.png", weight_t1_plot, base_aspect_ratio = 1)
# Subset to time point 2
weight_change_t1 <- weight_change[weight_change$time == 2, 1:5]
# How many are negative and how many are neutral/positive weight gain?
summary(weight_change_t1$Relapse_t3 >0) Mode FALSE TRUE NA's
logical 33 8 14 weight_change_t1[,6] <- weight_change_t1$Relapse_t3 >0
colnames(weight_change_t1) <- c("BAN_ID", "time", "Weight", "Weight_change", "Relapse_t3", "Relapse_notes")
weight_change_t1 <- weight_change_t1[complete.cases(weight_change_t1), ]
# Plot weight at T1
weight_t2_plot <- ggplot(weight_change_t1, aes(Relapse_notes, Weight)) + geom_boxplot() + xlab("Lost weight from T2 to T3") + ylab("Weight") + ggtitle("Weight at T2") + ylim(53,125)
save_plot("/Users/laurenblake/Dropbox/Figures/weight_t2.png", weight_t2_plot, base_aspect_ratio = 1)
# Weight change from T1 to T2
weight_change_t1 <- weight_change_t1[complete.cases(weight_change_t1), ]
weight_change_t1_plot <- ggplot(weight_change_t1, aes(Relapse_notes, Weight_change)) + geom_boxplot() + xlab("Lost weight from T2 to T3") + ylab("Weight change from T1 to T2") + ylim(-15,46)
plot_grid(weight_change_t1_plot)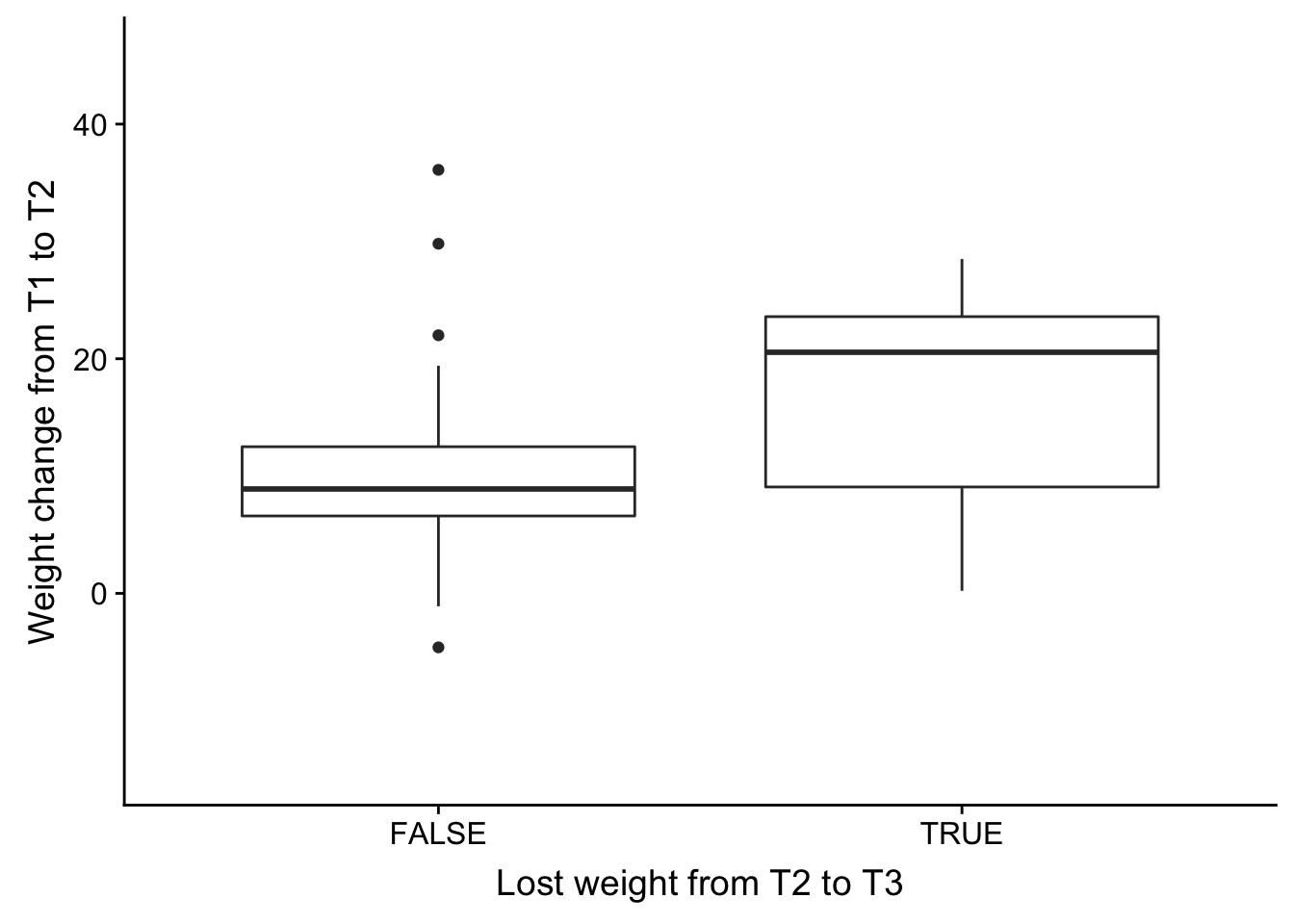
save_plot("/Users/laurenblake/Dropbox/Figures/weight_change_t1t2.png", weight_change_t1_plot, base_aspect_ratio = 1)
# Subset to time point 3
weight_change_t1 <- weight_change[weight_change$time == 3, 1:5]
# How many are negative and how many are neutral/positive weight gain?
summary(weight_change_t1$Relapse_t3 >0) Mode FALSE TRUE NA's
logical 33 8 14 weight_change_t1[,6] <- weight_change_t1$Relapse_t3 >0
colnames(weight_change_t1) <- c("BAN_ID", "time", "Weight", "Weight_change", "Relapse_t3", "Relapse_notes")
weight_change_t1 <- weight_change_t1[complete.cases(weight_change_t1), ]
# Plot weight at T1
weight_t3_plot <- ggplot(weight_change_t1, aes(Relapse_notes, Weight)) + geom_boxplot() + xlab("Lost weight from T2 to T3") + ylab("Weight") + ggtitle("Weight at T3") + ylim(53,125)
save_plot("/Users/laurenblake/Dropbox/Figures/weight_t3.png", weight_t3_plot, base_aspect_ratio = 1)Warning: Removed 6 rows containing non-finite values (stat_boxplot).weight_change_t1_plot <- ggplot(weight_change_t1, aes(Relapse_notes, Weight_change)) + geom_boxplot() + xlab("Lost weight from T2 to T3") + ylab("Weight change from T2 to T3") + ylim(-15,46)
plot_grid(weight_change_t1_plot)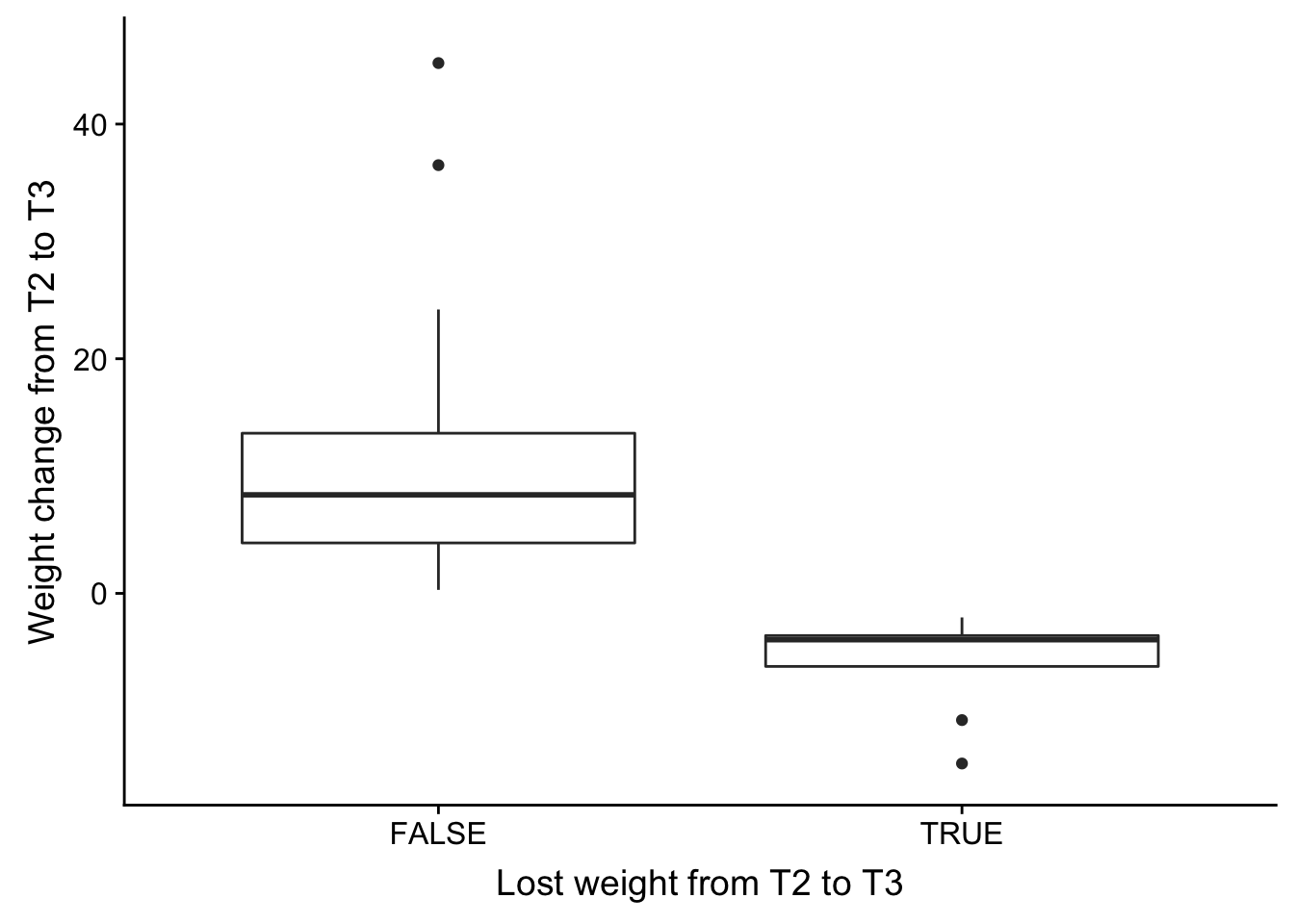
save_plot("/Users/laurenblake/Dropbox/Figures/weight_change_t2t3.png", weight_change_t1_plot, base_aspect_ratio = 1)Let’s look at the 8 individuals
# Let's look at the weight over time for the individuals with weight loss
weight_change_8 <- weight_change[weight_change$Relapse_t3 == 1,]
weight_change_8 <- weight_change_8[complete.cases(weight_change_8[,1:5]), ]
# Plot the individuals
ind_time <- ggplot(weight_change_8, aes(time, Weight, colour = as.factor(weight_change_8$BAN_ID))) + geom_point() + geom_line() + theme(legend.position="none") + xlab("Time")
plot_grid(ind_time)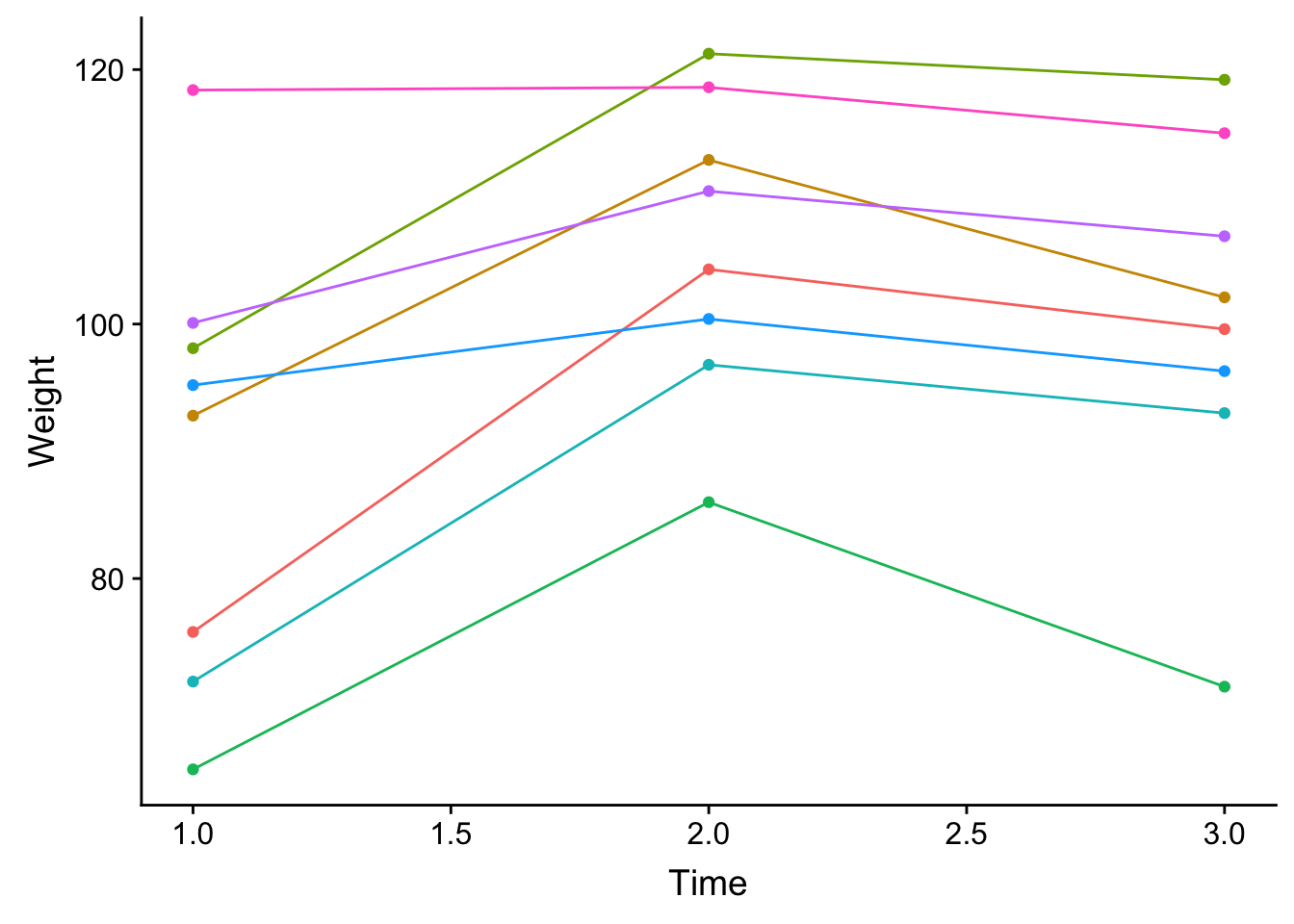
save_plot("/Users/laurenblake/Dropbox/Figures/weight_change_no_t3.png", ind_time, base_aspect_ratio = 1)
# Summarize the
weight_change_8_t3 <- weight_change_8[weight_change_8$time == 3,]
summary(weight_change_8_t3) BAN_ID time Weight Weight_change
Min. :2207 Min. :3 Min. : 71.50 Min. :-14.500
1st Qu.:2226 1st Qu.:3 1st Qu.: 95.47 1st Qu.: -6.225
Median :2238 Median :3 Median :100.85 Median : -3.950
Mean :2241 Mean :3 Mean :100.45 Mean : -5.889
3rd Qu.:2262 3rd Qu.:3 3rd Qu.:108.92 3rd Qu.: -3.595
Max. :2270 Max. :3 Max. :119.20 Max. : -2.050
Relapse_t3 Relapse_t4 Relapse_t5
Min. :1 Min. :0 Min. :0.00
1st Qu.:1 1st Qu.:0 1st Qu.:0.25
Median :1 Median :0 Median :0.50
Mean :1 Mean :0 Mean :0.50
3rd Qu.:1 3rd Qu.:0 3rd Qu.:0.75
Max. :1 Max. :0 Max. :1.00
NA's :6 NA's :6 Correlation between weight relapse and other variables
# Read in the clinical covariates
clinical_sample_info <- read.csv("../data/lm_covar_fixed_random.csv")
dim(clinical_sample_info)[1] 156 14# Subset to T1-T3
clinical_sample <- clinical_sample_info[1:144,(-12)]
dim(clinical_sample)[1] 144 13# Relapse
chisq.test(as.factor(clinical_sample$Time), as.factor(clinical_sample$Relapse_t3), simulate.p.value = TRUE)$p.value[1] 0.9585207# Individual-yes
chisq.test(as.factor(clinical_sample$Individ), as.factor(clinical_sample$Relapse_t3), simulate.p.value = TRUE)$p.value[1] 0.0004997501# Age-yes
summary(lm(clinical_sample$Age ~ as.factor(clinical_sample$Relapse_t3)))
Call:
lm(formula = clinical_sample$Age ~ as.factor(clinical_sample$Relapse_t3))
Residuals:
Min 1Q Median 3Q Max
-9.583 -5.583 -1.978 2.022 20.021
Coefficients:
Estimate Std. Error t value
(Intercept) 23.9785 0.8045 29.806
as.factor(clinical_sample$Relapse_t3)1 6.6048 1.7763 3.718
Pr(>|t|)
(Intercept) < 2e-16 ***
as.factor(clinical_sample$Relapse_t3)1 0.000311 ***
---
Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
Residual standard error: 7.758 on 115 degrees of freedom
(27 observations deleted due to missingness)
Multiple R-squared: 0.1073, Adjusted R-squared: 0.09956
F-statistic: 13.83 on 1 and 115 DF, p-value: 0.0003114# BE-no
chisq.test(as.factor(clinical_sample$BE_GROUP), as.factor(clinical_sample$Relapse_t3), simulate.p.value = TRUE)$p.value[1] 0.4742629# Psych meds- no
chisq.test(as.factor(clinical_sample$psychmeds), as.factor(clinical_sample$Relapse_t3), simulate.p.value = TRUE)$p.value[1] 0.8270865# RBC-no
summary(lm(clinical_sample$RBC ~ as.factor(clinical_sample$Relapse_t3)))
Call:
lm(formula = clinical_sample$RBC ~ as.factor(clinical_sample$Relapse_t3))
Residuals:
Min 1Q Median 3Q Max
-1.40750 -0.24763 0.02237 0.27237 1.22237
Coefficients:
Estimate Std. Error t value
(Intercept) 4.22763 0.04503 93.883
as.factor(clinical_sample$Relapse_t3)1 -0.01013 0.09943 -0.102
Pr(>|t|)
(Intercept) <2e-16 ***
as.factor(clinical_sample$Relapse_t3)1 0.919
---
Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
Residual standard error: 0.4343 on 115 degrees of freedom
(27 observations deleted due to missingness)
Multiple R-squared: 9.034e-05, Adjusted R-squared: -0.008605
F-statistic: 0.01039 on 1 and 115 DF, p-value: 0.919# AE-no
summary(lm(clinical_sample$AE ~ as.factor(clinical_sample$Relapse_t3)))
Call:
lm(formula = clinical_sample$AE ~ as.factor(clinical_sample$Relapse_t3))
Residuals:
Min 1Q Median 3Q Max
-0.25833 -0.06344 -0.06344 0.03656 2.64167
Coefficients:
Estimate Std. Error t value
(Intercept) 0.16344 0.02970 5.503
as.factor(clinical_sample$Relapse_t3)1 0.09489 0.06558 1.447
Pr(>|t|)
(Intercept) 2.29e-07 ***
as.factor(clinical_sample$Relapse_t3)1 0.151
---
Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
Residual standard error: 0.2864 on 115 degrees of freedom
(27 observations deleted due to missingness)
Multiple R-squared: 0.01788, Adjusted R-squared: 0.009343
F-statistic: 2.094 on 1 and 115 DF, p-value: 0.1506# Race- no
chisq.test(as.factor(clinical_sample$Race), as.factor(clinical_sample$Relapse_t3), simulate.p.value = TRUE)$p.value[1] 0.4582709# AL- no
summary(lm(clinical_sample$AL ~ as.factor(clinical_sample$Relapse_t3)))
Call:
lm(formula = clinical_sample$AL ~ as.factor(clinical_sample$Relapse_t3))
Residuals:
Min 1Q Median 3Q Max
-1.11505 -0.41505 -0.01505 0.48495 1.38495
Coefficients:
Estimate Std. Error t value
(Intercept) 1.81505 0.05936 30.580
as.factor(clinical_sample$Relapse_t3)1 -0.11505 0.13105 -0.878
Pr(>|t|)
(Intercept) <2e-16 ***
as.factor(clinical_sample$Relapse_t3)1 0.382
---
Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
Residual standard error: 0.5724 on 115 degrees of freedom
(27 observations deleted due to missingness)
Multiple R-squared: 0.006657, Adjusted R-squared: -0.00198
F-statistic: 0.7707 on 1 and 115 DF, p-value: 0.3818# AN- no
summary(lm(clinical_sample$AN ~ as.factor(clinical_sample$Relapse_t3)))
Call:
lm(formula = clinical_sample$AN ~ as.factor(clinical_sample$Relapse_t3))
Residuals:
Min 1Q Median 3Q Max
-2.2258 -0.9258 -0.3258 0.5742 4.5500
Coefficients:
Estimate Std. Error t value
(Intercept) 3.0258 0.1282 23.607
as.factor(clinical_sample$Relapse_t3)1 0.2242 0.2830 0.792
Pr(>|t|)
(Intercept) <2e-16 ***
as.factor(clinical_sample$Relapse_t3)1 0.43
---
Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
Residual standard error: 1.236 on 115 degrees of freedom
(27 observations deleted due to missingness)
Multiple R-squared: 0.005428, Adjusted R-squared: -0.003221
F-statistic: 0.6276 on 1 and 115 DF, p-value: 0.4299# RIN- no
summary(lm(clinical_sample$RIN ~ as.factor(clinical_sample$Relapse_t3)))
Call:
lm(formula = clinical_sample$RIN ~ as.factor(clinical_sample$Relapse_t3))
Residuals:
Min 1Q Median 3Q Max
-2.37742 -0.27742 0.02258 0.42258 1.22258
Coefficients:
Estimate Std. Error t value
(Intercept) 6.67742 0.07043 94.809
as.factor(clinical_sample$Relapse_t3)1 -0.10659 0.15551 -0.685
Pr(>|t|)
(Intercept) <2e-16 ***
as.factor(clinical_sample$Relapse_t3)1 0.494
---
Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
Residual standard error: 0.6792 on 115 degrees of freedom
(27 observations deleted due to missingness)
Multiple R-squared: 0.004069, Adjusted R-squared: -0.004592
F-statistic: 0.4698 on 1 and 115 DF, p-value: 0.4945Session information
sessionInfo()R version 3.4.3 (2017-11-30)
Platform: x86_64-apple-darwin15.6.0 (64-bit)
Running under: OS X El Capitan 10.11.6
Matrix products: default
BLAS: /Library/Frameworks/R.framework/Versions/3.4/Resources/lib/libRblas.0.dylib
LAPACK: /Library/Frameworks/R.framework/Versions/3.4/Resources/lib/libRlapack.dylib
locale:
[1] en_US.UTF-8/en_US.UTF-8/en_US.UTF-8/C/en_US.UTF-8/en_US.UTF-8
attached base packages:
[1] stats graphics grDevices utils datasets methods base
other attached packages:
[1] cowplot_0.9.3 ggplot2_3.0.0
loaded via a namespace (and not attached):
[1] Rcpp_0.12.18 compiler_3.4.3 pillar_1.3.0
[4] git2r_0.23.0 plyr_1.8.4 workflowr_1.1.1
[7] bindr_0.1.1 R.methodsS3_1.7.1 R.utils_2.7.0
[10] tools_3.4.3 digest_0.6.16 evaluate_0.11
[13] tibble_1.4.2 gtable_0.2.0 pkgconfig_2.0.2
[16] rlang_0.2.2 yaml_2.2.0 bindrcpp_0.2.2
[19] withr_2.1.2 stringr_1.3.1 dplyr_0.7.6
[22] knitr_1.20 rprojroot_1.3-2 grid_3.4.3
[25] tidyselect_0.2.4 glue_1.3.0 R6_2.2.2
[28] rmarkdown_1.10 purrr_0.2.5 magrittr_1.5
[31] whisker_0.3-2 backports_1.1.2 scales_1.0.0
[34] htmltools_0.3.6 assertthat_0.2.0 colorspace_1.3-2
[37] labeling_0.3 stringi_1.2.4 lazyeval_0.2.1
[40] munsell_0.5.0 crayon_1.3.4 R.oo_1.22.0
This reproducible R Markdown analysis was created with workflowr 1.1.1