voom_limma_hg37
Lauren Blake
2018-08-28
Last updated: 2018-11-15
workflowr checks: (Click a bullet for more information)-
✔ R Markdown file: up-to-date
Great! Since the R Markdown file has been committed to the Git repository, you know the exact version of the code that produced these results.
-
✔ Environment: empty
Great job! The global environment was empty. Objects defined in the global environment can affect the analysis in your R Markdown file in unknown ways. For reproduciblity it’s best to always run the code in an empty environment.
-
✔ Seed:
set.seed(12345)The command
set.seed(12345)was run prior to running the code in the R Markdown file. Setting a seed ensures that any results that rely on randomness, e.g. subsampling or permutations, are reproducible. -
✔ Session information: recorded
Great job! Recording the operating system, R version, and package versions is critical for reproducibility.
-
Great! You are using Git for version control. Tracking code development and connecting the code version to the results is critical for reproducibility. The version displayed above was the version of the Git repository at the time these results were generated.✔ Repository version: 7bcd123
Note that you need to be careful to ensure that all relevant files for the analysis have been committed to Git prior to generating the results (you can usewflow_publishorwflow_git_commit). workflowr only checks the R Markdown file, but you know if there are other scripts or data files that it depends on. Below is the status of the Git repository when the results were generated:
Note that any generated files, e.g. HTML, png, CSS, etc., are not included in this status report because it is ok for generated content to have uncommitted changes.Ignored files: Ignored: .DS_Store Ignored: analysis/.DS_Store Ignored: analysis/VennDiagram2018-07-24_06-55-46.log Ignored: analysis/VennDiagram2018-07-24_06-56-13.log Ignored: analysis/VennDiagram2018-07-24_06-56-50.log Ignored: analysis/VennDiagram2018-07-24_06-58-41.log Ignored: analysis/VennDiagram2018-07-24_07-00-07.log Ignored: analysis/VennDiagram2018-07-24_07-00-42.log Ignored: analysis/VennDiagram2018-07-24_07-01-08.log Ignored: analysis/VennDiagram2018-08-17_15-13-24.log Ignored: analysis/VennDiagram2018-08-17_15-13-30.log Ignored: analysis/VennDiagram2018-08-17_15-15-06.log Ignored: analysis/VennDiagram2018-08-17_15-16-01.log Ignored: analysis/VennDiagram2018-08-17_15-17-51.log Ignored: analysis/VennDiagram2018-08-17_15-18-42.log Ignored: analysis/VennDiagram2018-08-17_15-19-21.log Ignored: analysis/VennDiagram2018-08-20_09-07-57.log Ignored: analysis/VennDiagram2018-08-20_09-08-37.log Ignored: analysis/VennDiagram2018-08-26_19-54-03.log Ignored: analysis/VennDiagram2018-08-26_20-47-08.log Ignored: analysis/VennDiagram2018-08-26_20-49-49.log Ignored: analysis/VennDiagram2018-08-27_00-04-36.log Ignored: analysis/VennDiagram2018-08-27_00-09-27.log Ignored: analysis/VennDiagram2018-08-27_00-13-57.log Ignored: analysis/VennDiagram2018-08-27_00-16-32.log Ignored: analysis/VennDiagram2018-08-27_10-00-25.log Ignored: analysis/VennDiagram2018-08-28_06-03-13.log Ignored: analysis/VennDiagram2018-08-28_06-03-14.log Ignored: analysis/VennDiagram2018-08-28_06-05-50.log Ignored: analysis/VennDiagram2018-08-28_06-06-58.log Ignored: analysis/VennDiagram2018-08-28_06-10-12.log Ignored: analysis/VennDiagram2018-08-28_06-10-13.log Ignored: analysis/VennDiagram2018-08-28_06-18-29.log Ignored: analysis/VennDiagram2018-08-28_07-22-26.log Ignored: analysis/VennDiagram2018-08-28_07-22-27.log Ignored: analysis/VennDiagram2018-08-28_13-05-27.log Ignored: analysis/VennDiagram2018-09-12_01-45-59.log Ignored: analysis/VennDiagram2018-09-12_01-49-31.log Ignored: analysis/VennDiagram2018-09-12_01-58-11.log Ignored: analysis/VennDiagram2018-09-12_01-59-46.log Ignored: analysis/VennDiagram2018-09-12_02-08-07.log Ignored: analysis/VennDiagram2018-09-12_02-08-56.log Ignored: analysis/VennDiagram2018-11-15_14-20-08.log Ignored: analysis/VennDiagram2018-11-15_14-20-15.log Ignored: analysis/VennDiagram2018-11-15_14-20-23.log Ignored: analysis/VennDiagram2018-11-15_14-21-14.log Ignored: analysis/VennDiagram2018-11-15_14-21-57.log Ignored: analysis/VennDiagram2018-11-15_14-33-34.log Ignored: analysis/VennDiagram2018-11-15_14-36-19.log Ignored: analysis/VennDiagram2018-11-15_14-48-41.log Ignored: analysis/VennDiagram2018-11-15_14-48-42.log Ignored: analysis/VennDiagram2018-11-15_15-03-35.log Ignored: analysis/VennDiagram2018-11-15_15-03-55.log Ignored: analysis/VennDiagram2018-11-15_15-07-05.log Ignored: analysis/VennDiagram2018-11-15_15-07-25.log Ignored: analysis/VennDiagram2018-11-15_15-09-29.log Ignored: analysis/VennDiagram2018-11-15_15-09-48.log Ignored: analysis/VennDiagram2018-11-15_15-14-30.log Ignored: data/DAVID_2covar/ Ignored: data/DAVID_results/ Ignored: data/Eigengenes/ Ignored: data/aux_info/ Ignored: data/hg_38/ Ignored: data/libParams/ Ignored: data/logs/ Ignored: docs/VennDiagram2018-07-24_06-55-46.log Ignored: docs/VennDiagram2018-07-24_06-56-13.log Ignored: docs/VennDiagram2018-07-24_06-56-50.log Ignored: docs/VennDiagram2018-07-24_06-58-41.log Ignored: docs/VennDiagram2018-07-24_07-00-07.log Ignored: docs/VennDiagram2018-07-24_07-00-42.log Ignored: docs/VennDiagram2018-07-24_07-01-08.log Ignored: docs/figure/.DS_Store Ignored: output/.DS_Store Untracked files: Untracked: docs/figure/time_two_covar.Rmd/ Unstaged changes: Modified: analysis/time_two_covar.Rmd
Expand here to see past versions:
Introduction
# Library
library(edgeR)Loading required package: limmalibrary(limma)
library(VennDiagram)Warning: package 'VennDiagram' was built under R version 3.4.4Loading required package: gridLoading required package: futile.loggerlibrary(cowplot)Warning: package 'cowplot' was built under R version 3.4.4Loading required package: ggplot2Warning: package 'ggplot2' was built under R version 3.4.4
Attaching package: 'cowplot'The following object is masked from 'package:ggplot2':
ggsave# Read in the data
tx.salmon <- readRDS("../data/counts_hg37_gc_txsalmon.RData")
salmon_counts<- as.data.frame(tx.salmon$counts)
#tx.salmon <- readRDS("../data/counts_hg38_gc_dds.RData")
#salmon_counts<- as.data.frame(tx.salmon)
# Subset to T1-T3
salmon_counts <- salmon_counts[,1:144]
# Read in the clinical covariates
clinical_sample_info <- read.csv("../data/lm_covar_fixed_random.csv")
dim(clinical_sample_info)[1] 156 14# Subset to T1-T3
clinical_sample <- clinical_sample_info[1:144,(-12)]
dim(clinical_sample)[1] 144 13Differential expression pipeline
# Filter lowly expressed reads
cpm <- cpm(salmon_counts, log=TRUE)
expr_cutoff <- 1.5
hist(cpm, main = "log2(CPM) values in unfiltered data", breaks = 100, xlab = "log2(CPM) values")
abline(v = expr_cutoff, col = "red", lwd = 3)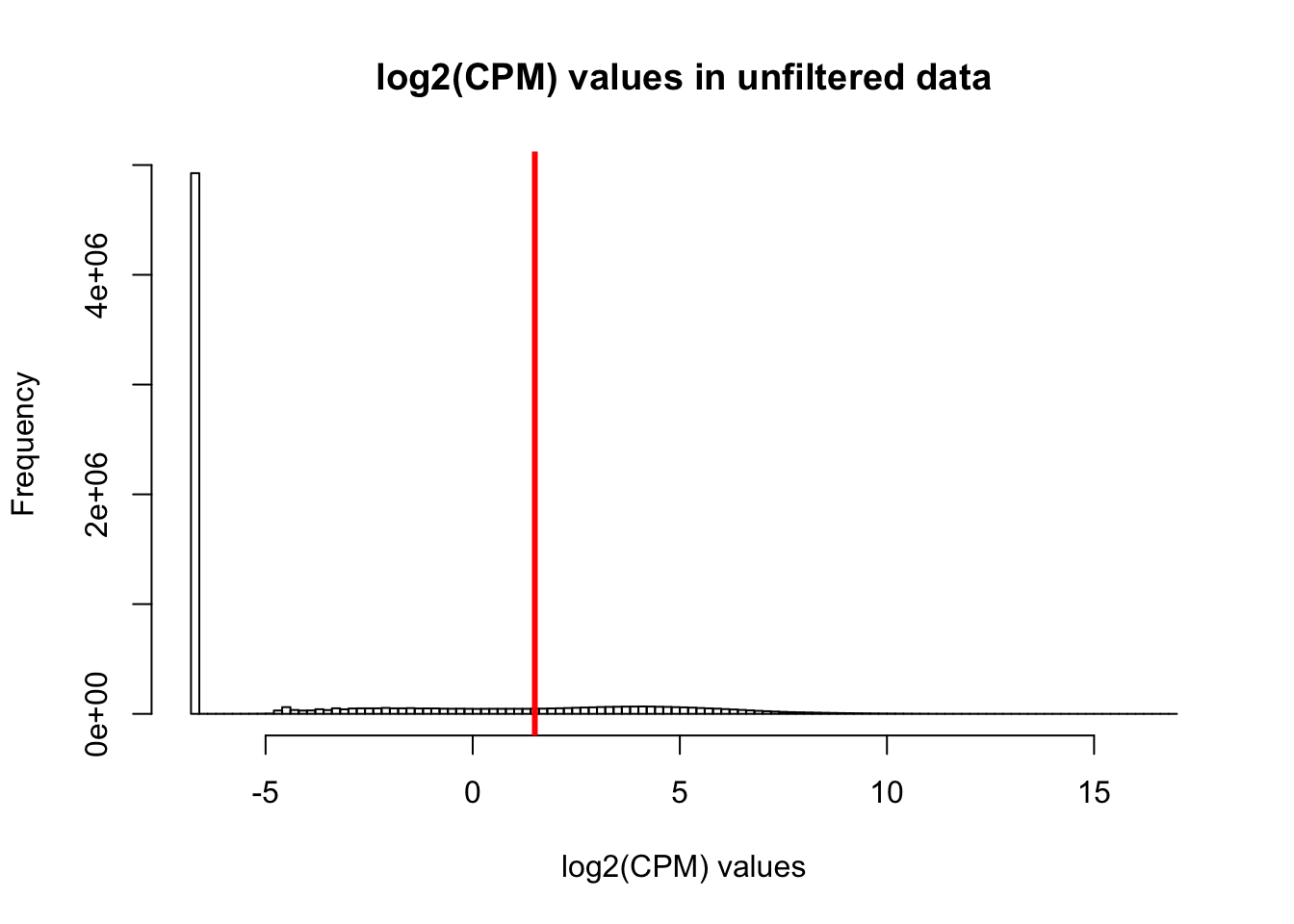
Expand here to see past versions of unnamed-chunk-2-1.png:
| Version | Author | Date |
|---|---|---|
| 7bcd123 | Lauren Blake | 2018-11-15 |
hist(cpm, main = "log2(CPM) values in unfiltered data", breaks = 100, xlab = "log2(CPM) values", ylim = c(0, 100000))
abline(v = expr_cutoff, col = "red", lwd = 3)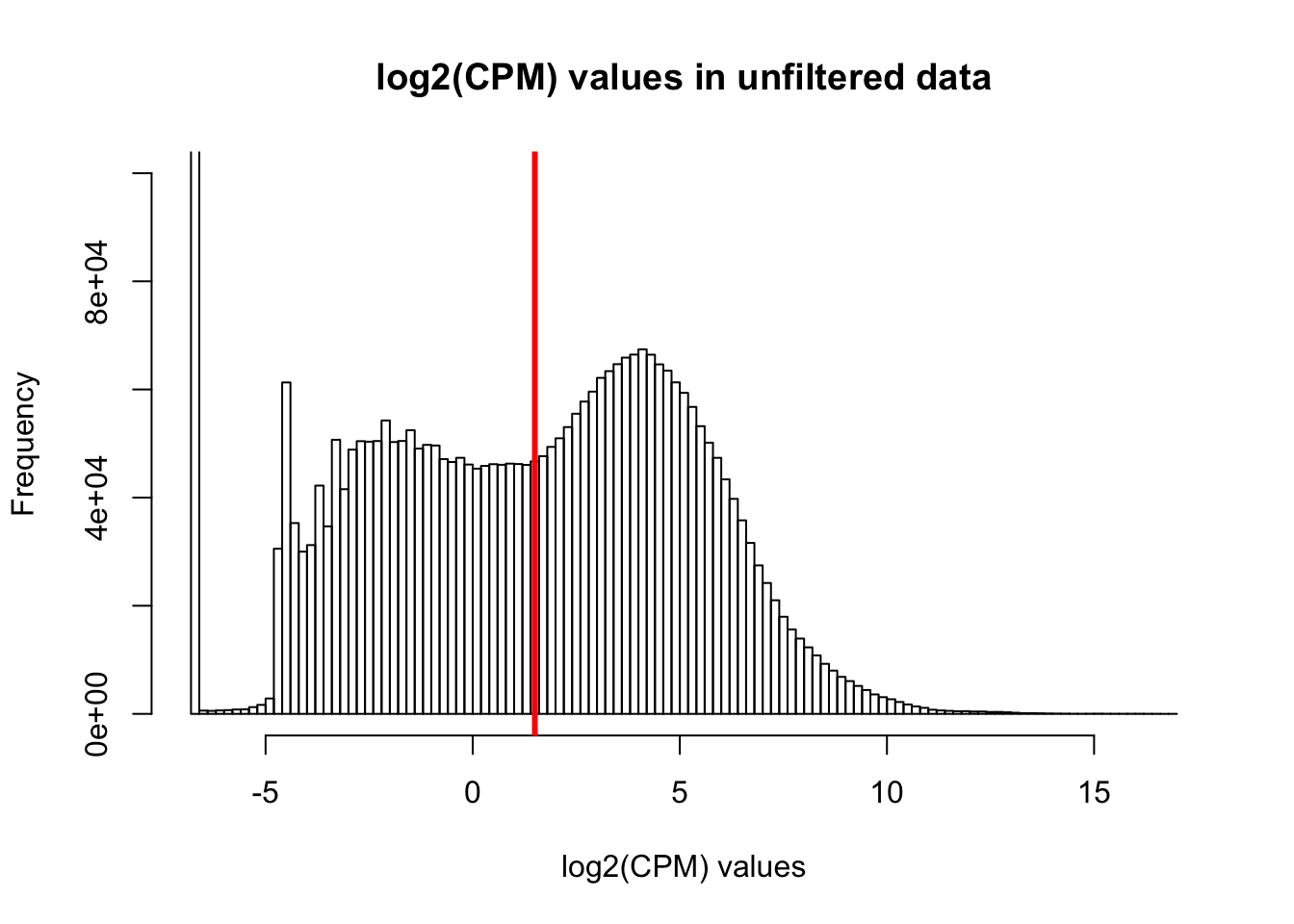
Expand here to see past versions of unnamed-chunk-2-2.png:
| Version | Author | Date |
|---|---|---|
| 7bcd123 | Lauren Blake | 2018-11-15 |
# Basic filtering
cpm_filtered <- (rowSums(cpm > 1.5) > 72)
genes_in_cutoff <- cpm[cpm_filtered==TRUE,]
hist(as.numeric(unlist(genes_in_cutoff)), main = "log2(CPM) values in filtered data", breaks = 100, xlab = "log2(CPM) values")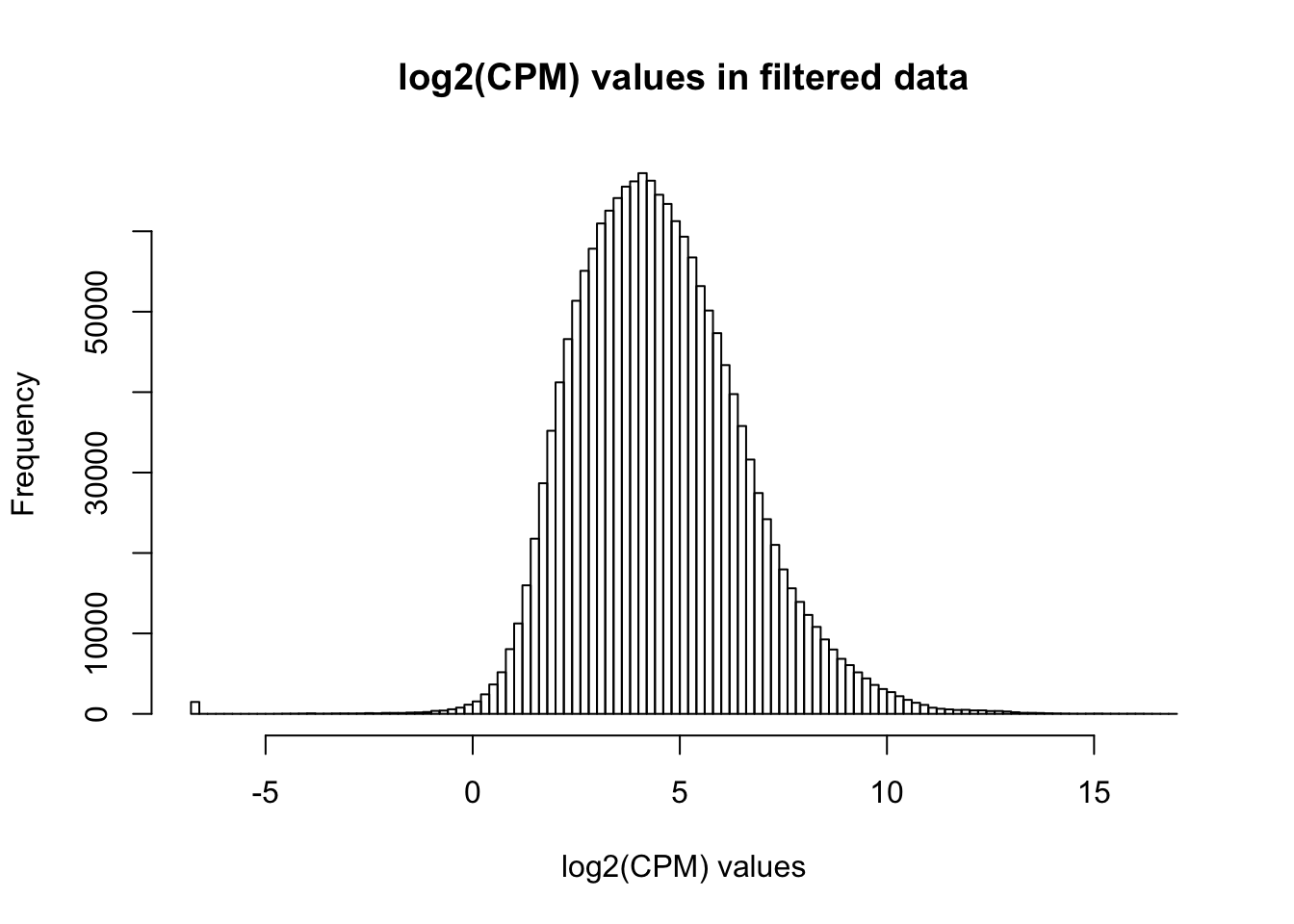
Expand here to see past versions of unnamed-chunk-2-3.png:
| Version | Author | Date |
|---|---|---|
| 7bcd123 | Lauren Blake | 2018-11-15 |
# Find the original counts of all of the genes that fit the criteria
counts_genes_in_cutoff <- salmon_counts[cpm_filtered==TRUE,]
dim(counts_genes_in_cutoff)[1] 11501 144# Filter out hemoglobin
counts_genes_in_cutoff <- counts_genes_in_cutoff[which( rownames(counts_genes_in_cutoff) != "HBB" ),]
counts_genes_in_cutoff <- counts_genes_in_cutoff[which( rownames(counts_genes_in_cutoff) != "HBA2" ),]
counts_genes_in_cutoff <- counts_genes_in_cutoff[which( rownames(counts_genes_in_cutoff) != "HBA1" ),]
# Take the TMM of the counts only for the genes that remain after filtering
dge_in_cutoff <- DGEList(counts=as.matrix(counts_genes_in_cutoff), genes=rownames(counts_genes_in_cutoff), group = as.character(t(clinical_sample$Individual)))
dge_in_cutoff <- calcNormFactors(dge_in_cutoff)
cpm_in_cutoff <- cpm(dge_in_cutoff, normalized.lib.sizes=TRUE, log=TRUE)
pca_genes <- prcomp(t(cpm_in_cutoff), scale = T, retx = TRUE, center = TRUE)
matrixpca <- pca_genes$x
PC1 <- matrixpca[,1]
PC2 <- matrixpca[,2]
pc3 <- matrixpca[,3]
pc4 <- matrixpca[,4]
pc5 <- matrixpca[,5]
pcs <- data.frame(PC1, PC2, pc3, pc4, pc5)
summary <- summary(pca_genes)
head(summary$importance[2,1:5]) PC1 PC2 PC3 PC4 PC5
0.25084 0.12998 0.08768 0.05670 0.03298 norm_count <- ggplot(data=pcs, aes(x=PC1, y=PC2, color= as.factor(clinical_sample$Time))) + geom_point(aes(colour = as.factor(clinical_sample$Time))) + ggtitle("PCA of normalized counts") + scale_color_discrete(name = "Time")
plot_grid(norm_count)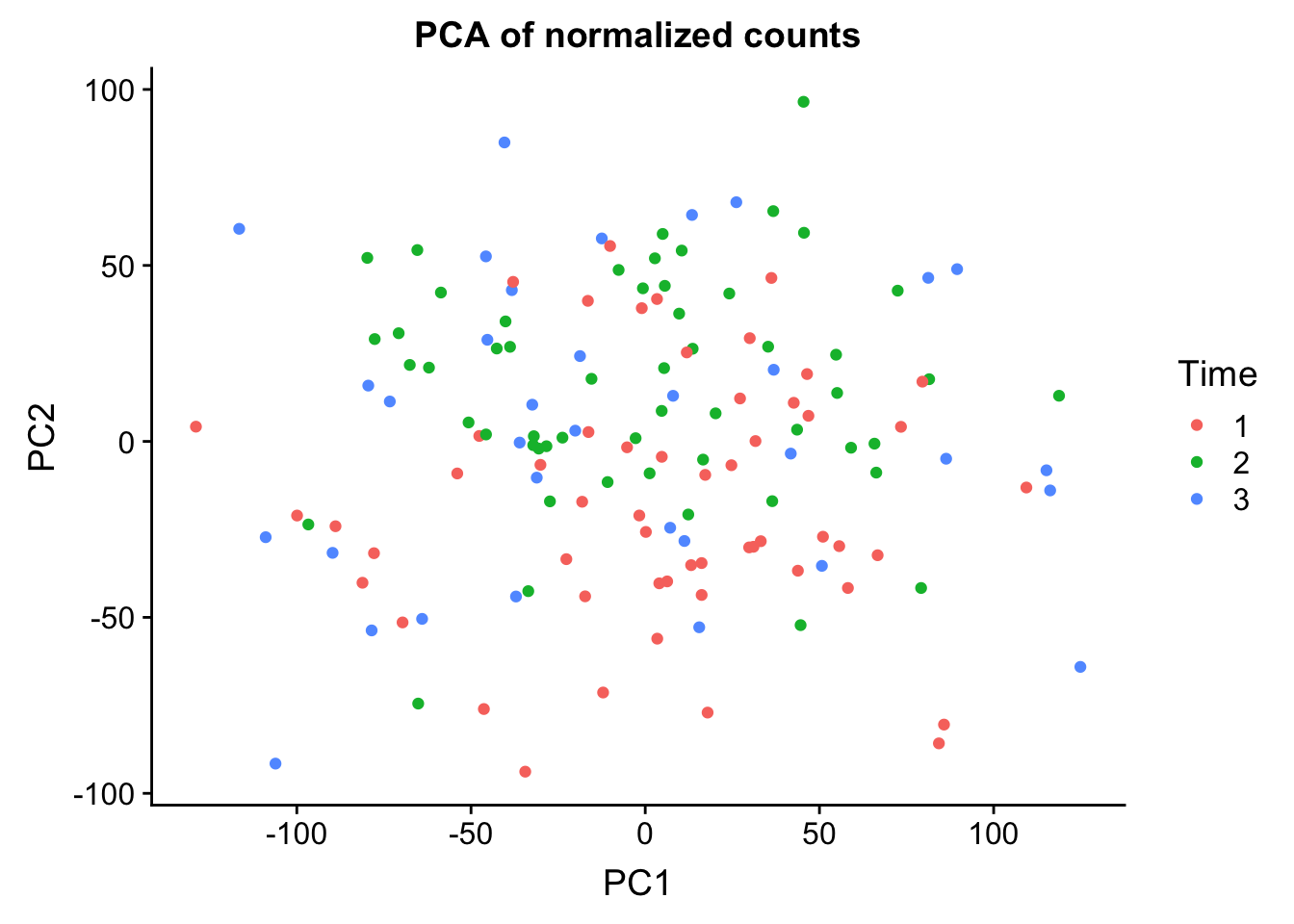
Expand here to see past versions of unnamed-chunk-2-4.png:
| Version | Author | Date |
|---|---|---|
| 7bcd123 | Lauren Blake | 2018-11-15 |
clinical_sample[,1] <- as.factor(clinical_sample[,1])
clinical_sample[,2] <- as.factor(clinical_sample[,2])
clinical_sample[,4] <- as.factor(clinical_sample[,4])
clinical_sample[,5] <- as.factor(clinical_sample[,5])
clinical_sample[,6] <- as.factor(clinical_sample[,6])
# Create the design matrix
# Use the standard treatment-contrasts parametrization. See Ch. 9 of limma
# User's Guide.
design <- model.matrix(~as.factor(Time) + Age + as.factor(Race) + as.factor(BE_GROUP) + as.factor(psychmeds) + RBC + AN + AE + AL + RIN, data = clinical_sample)
colnames(design) <- c("Intercept", "Time2", "Time3", "Race3", "Race5", "Age", "BE", "Psychmeds", "RBC", "AN", "AE", "AL", "RIN")
# Fit model
# Model individual as a random effect.
# Recommended to run both voom and duplicateCorrelation twice.
# https://support.bioconductor.org/p/59700/#67620
cpm.voom <- voom(dge_in_cutoff, design, normalize.method="none")
#check_rel <- duplicateCorrelation(cpm.voom, design, block = clinical_sample$Individual)
check_rel_correlation <- 0.1179835
cpm.voom.corfit <- voom(dge_in_cutoff, design, normalize.method="none", plot = TRUE, block = clinical_sample$Individual, correlation = check_rel_correlation)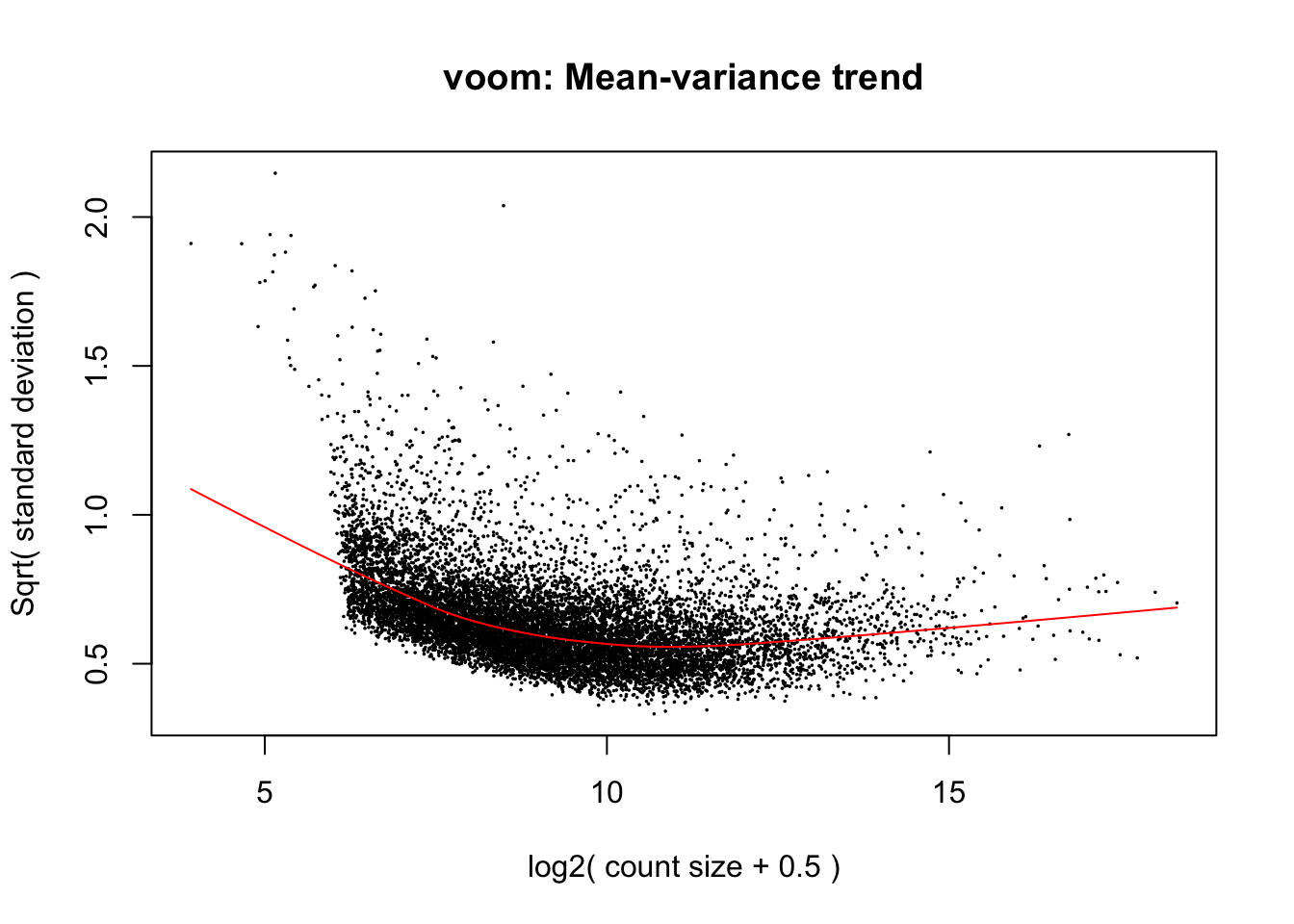
Expand here to see past versions of unnamed-chunk-2-5.png:
| Version | Author | Date |
|---|---|---|
| 7bcd123 | Lauren Blake | 2018-11-15 |
#check_rel <- duplicateCorrelation(cpm.voom.corfit, design, block = clinical_sample$Individual)
check_rel_correlation <- 0.1188083
plotDensities(cpm.voom.corfit[,1])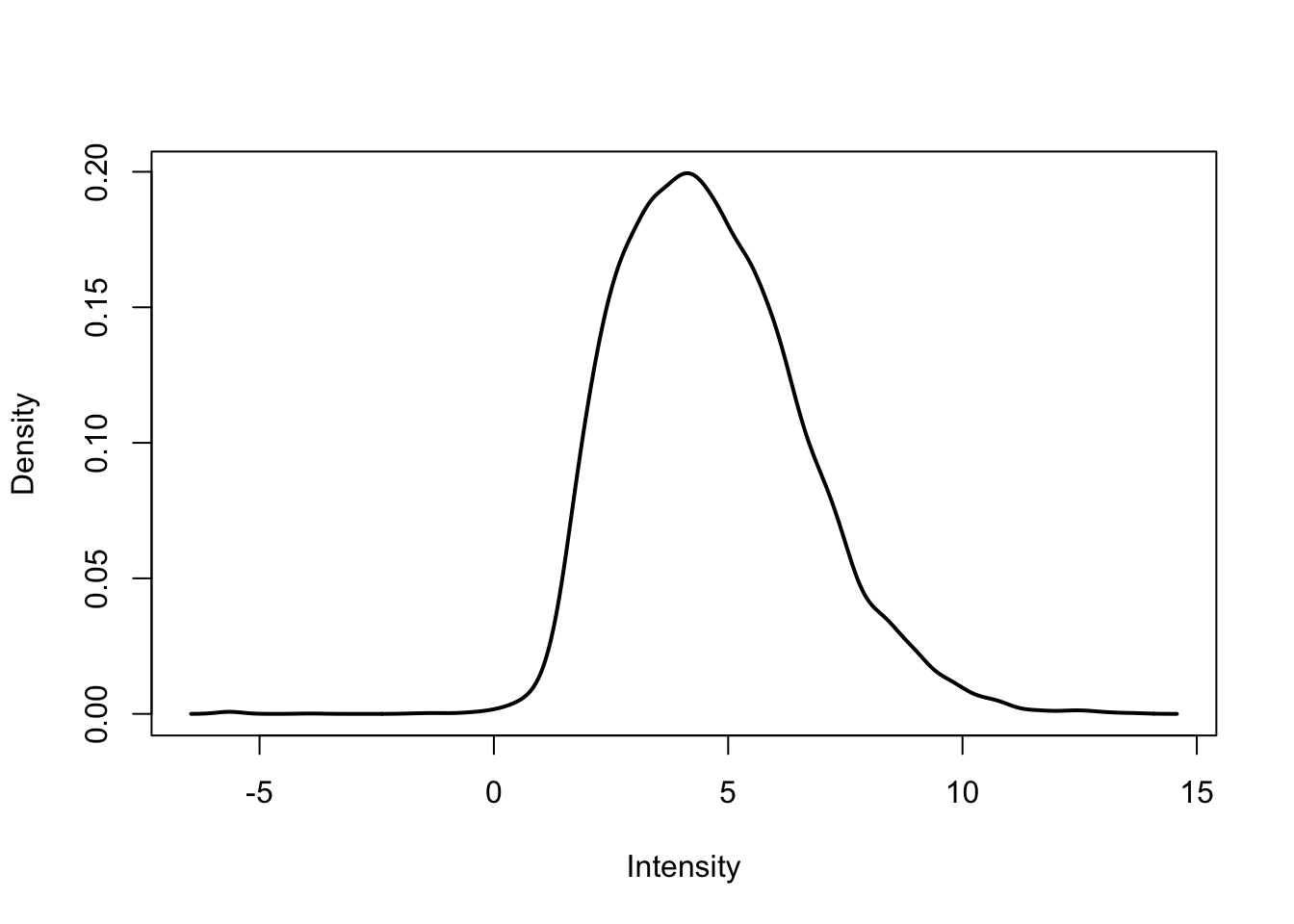
Expand here to see past versions of unnamed-chunk-2-6.png:
| Version | Author | Date |
|---|---|---|
| 7bcd123 | Lauren Blake | 2018-11-15 |
plotDensities(cpm.voom.corfit[,2])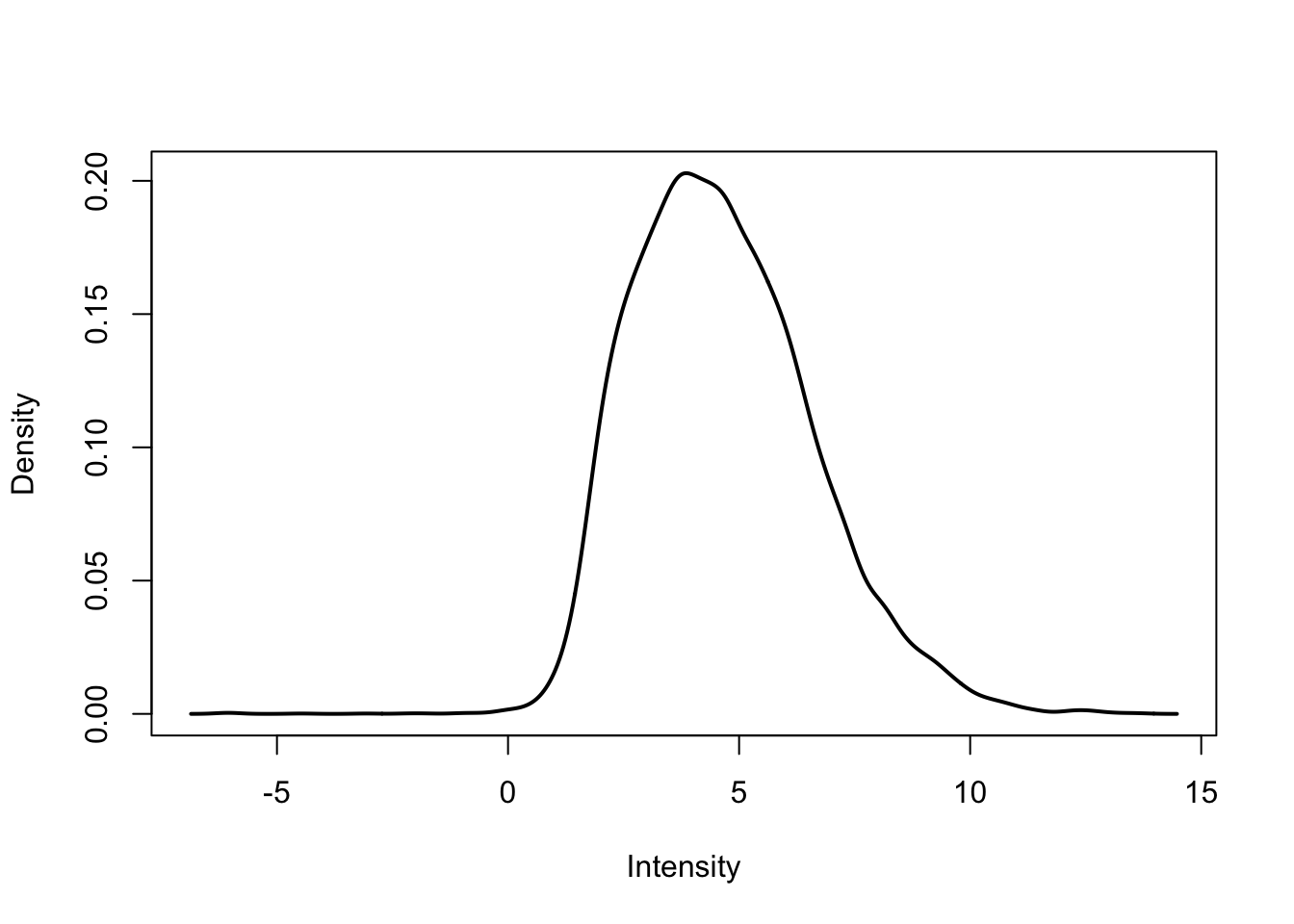
Expand here to see past versions of unnamed-chunk-2-7.png:
| Version | Author | Date |
|---|---|---|
| 7bcd123 | Lauren Blake | 2018-11-15 |
plotDensities(cpm.voom.corfit[,3])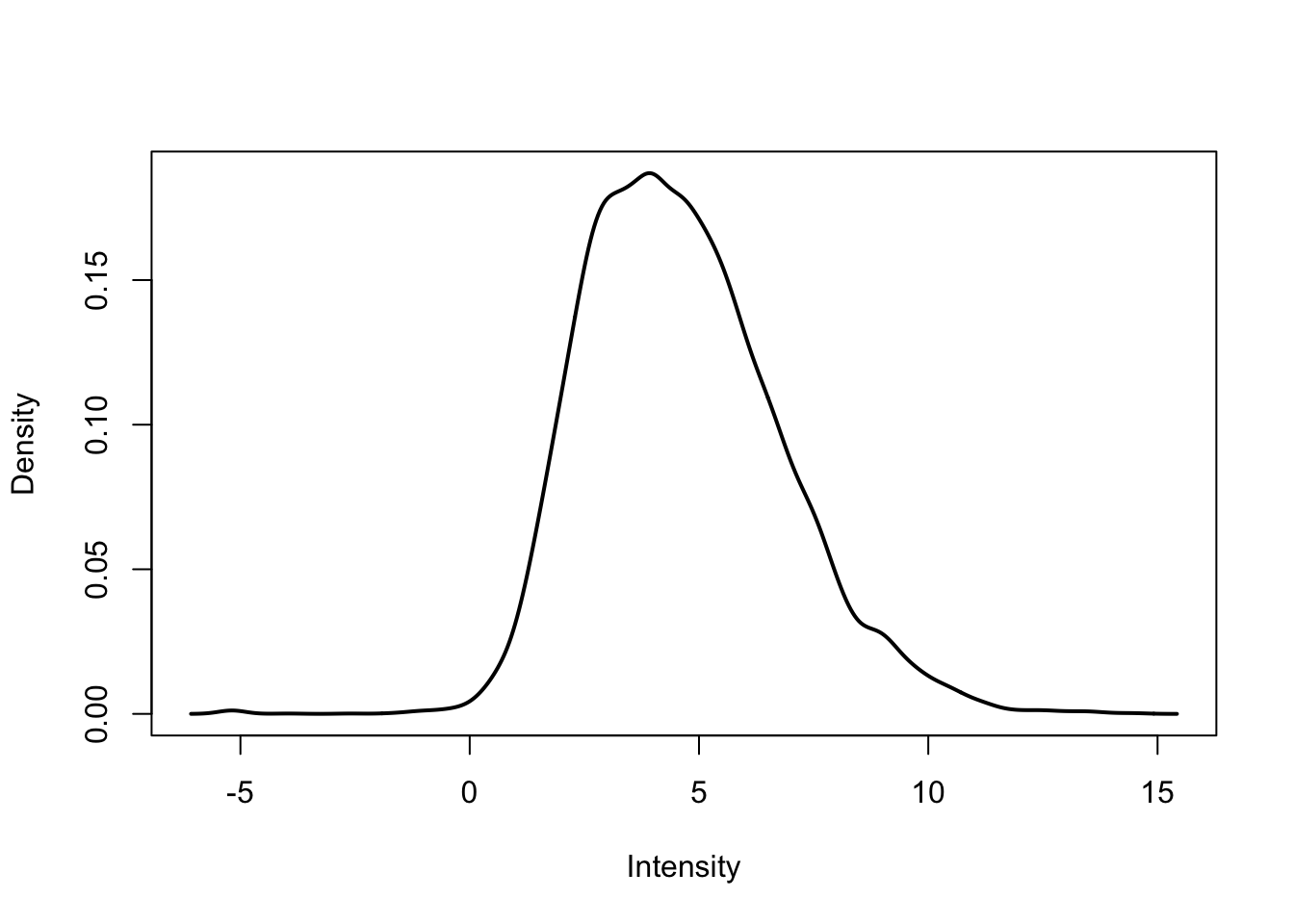
Expand here to see past versions of unnamed-chunk-2-8.png:
| Version | Author | Date |
|---|---|---|
| 7bcd123 | Lauren Blake | 2018-11-15 |
pca_genes <- prcomp(t(cpm.voom.corfit$E), scale = T, retx = TRUE, center = TRUE)
matrixpca <- pca_genes$x
PC1 <- matrixpca[,1]
PC2 <- matrixpca[,2]
pc3 <- matrixpca[,3]
pc4 <- matrixpca[,4]
pc5 <- matrixpca[,5]
pcs <- data.frame(PC1, PC2, pc3, pc4, pc5)
summary <- summary(pca_genes)
head(summary$importance[2,1:5]) PC1 PC2 PC3 PC4 PC5
0.25138 0.13024 0.08787 0.05642 0.03303 ggplot(data=pcs, aes(x=PC1, y=PC2, color=clinical_sample$Time)) + geom_point(aes(colour = as.factor(clinical_sample$Time))) + ggtitle("PCA of normalized counts") + scale_color_discrete(name = "Time")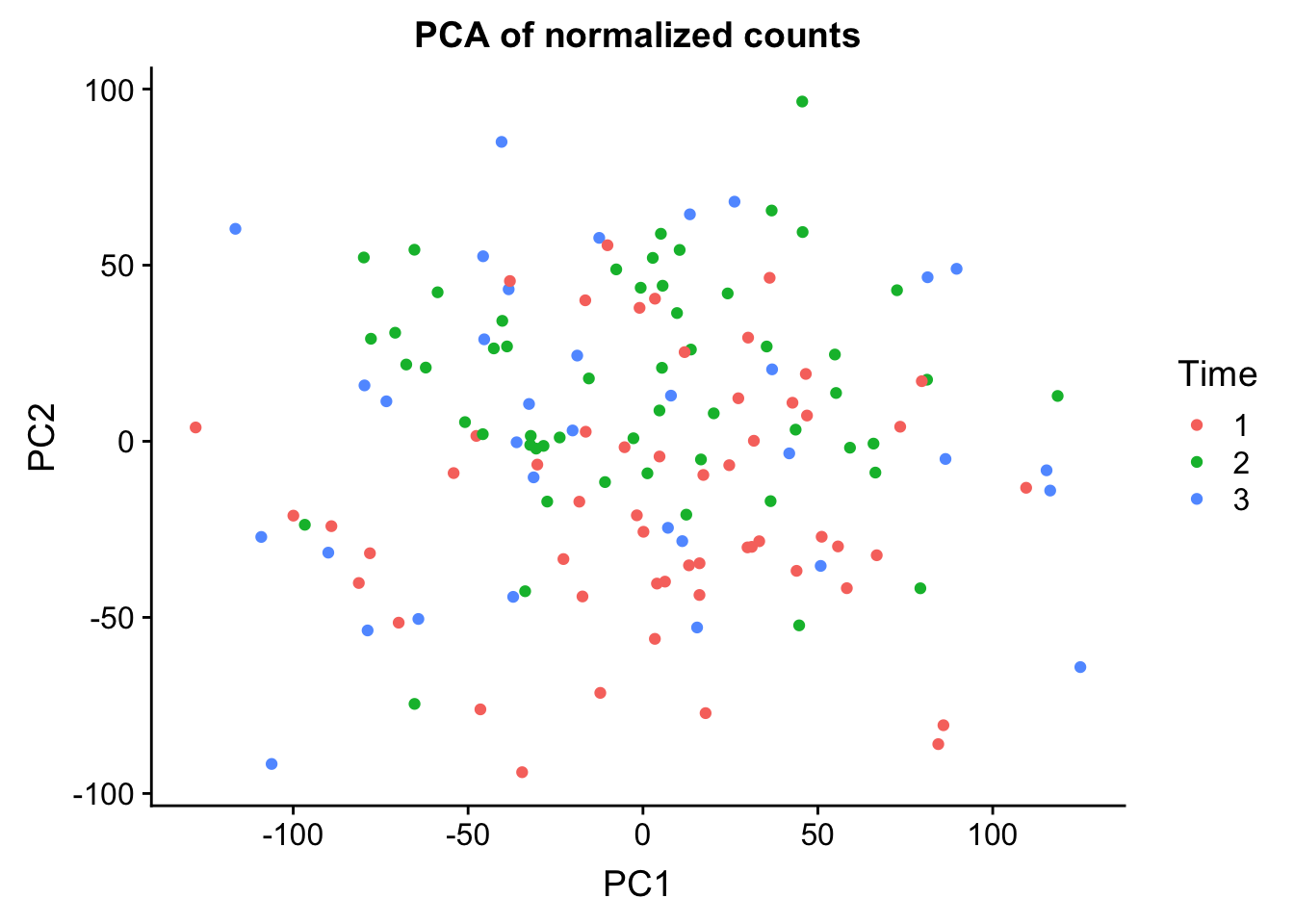
Expand here to see past versions of unnamed-chunk-2-9.png:
| Version | Author | Date |
|---|---|---|
| 7bcd123 | Lauren Blake | 2018-11-15 |
# Run lmFit and eBayes in limma
fit <- lmFit(cpm.voom.corfit, design, block=clinical_sample$Individual, correlation=check_rel_correlation)
# In the contrast matrix, have the time points
cm1 <- makeContrasts(Time1v2 = Time2, Time2v3 = Time3 - Time2, levels = design)
#cm1 <- makeContrasts(Time1v2 = Time2, Time2v3 = Time3, levels = design)
# Fit the new model
diff_species <- contrasts.fit(fit, cm1)
fit1 <- eBayes(diff_species)
FDR_level <- 0.05
Time1v2 =topTable(fit1, coef=1, adjust="BH", number=Inf, sort.by="none")
Time2v3 =topTable(fit1, coef=2, adjust="BH", number=Inf, sort.by="none")
#plot(fit1$coefficients[,1], fit1$coefficients[,2])
plot(Time1v2$logFC, Time2v3$logFC)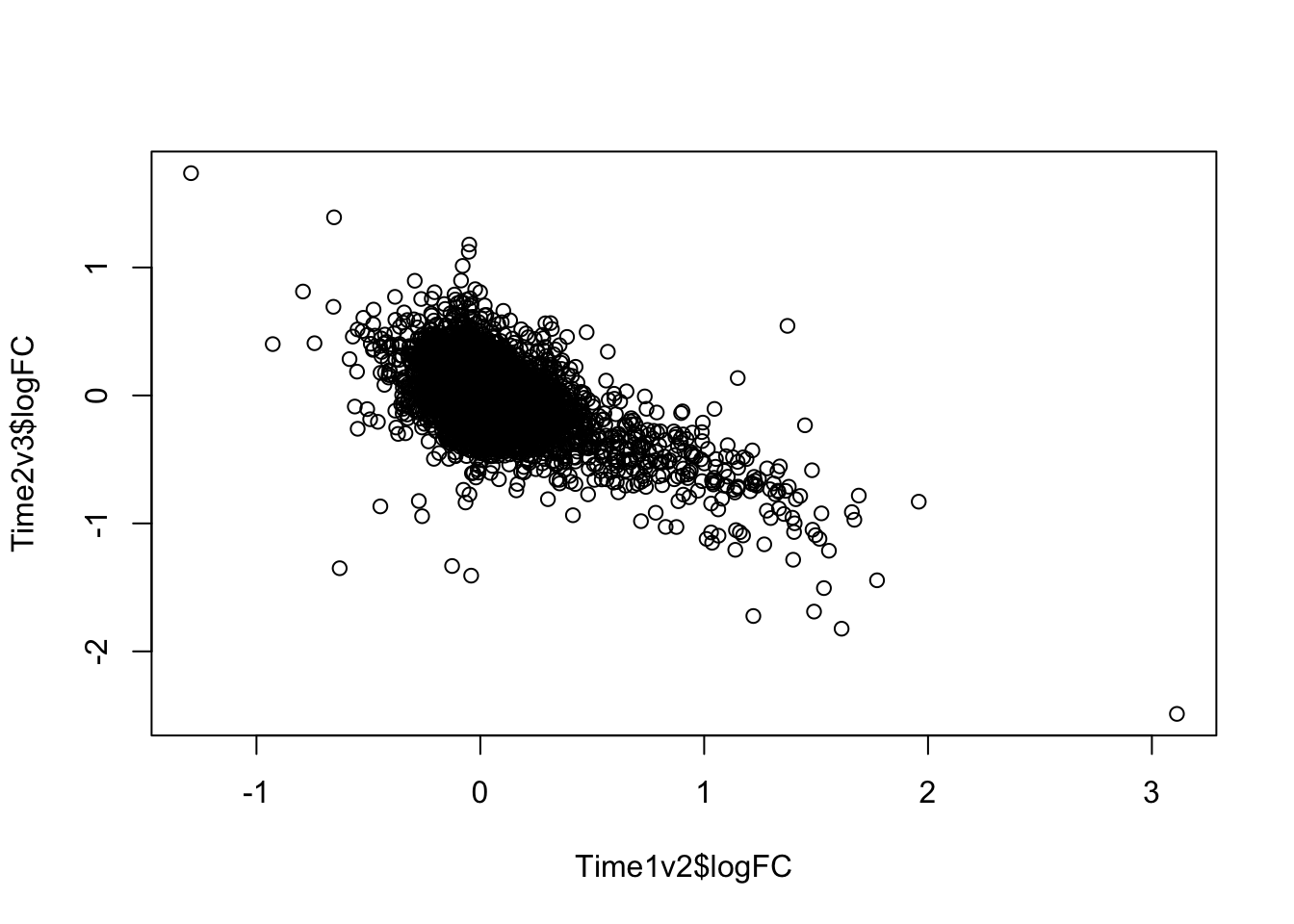
Expand here to see past versions of unnamed-chunk-2-10.png:
| Version | Author | Date |
|---|---|---|
| 7bcd123 | Lauren Blake | 2018-11-15 |
plot(Time1v2$t, Time2v3$t)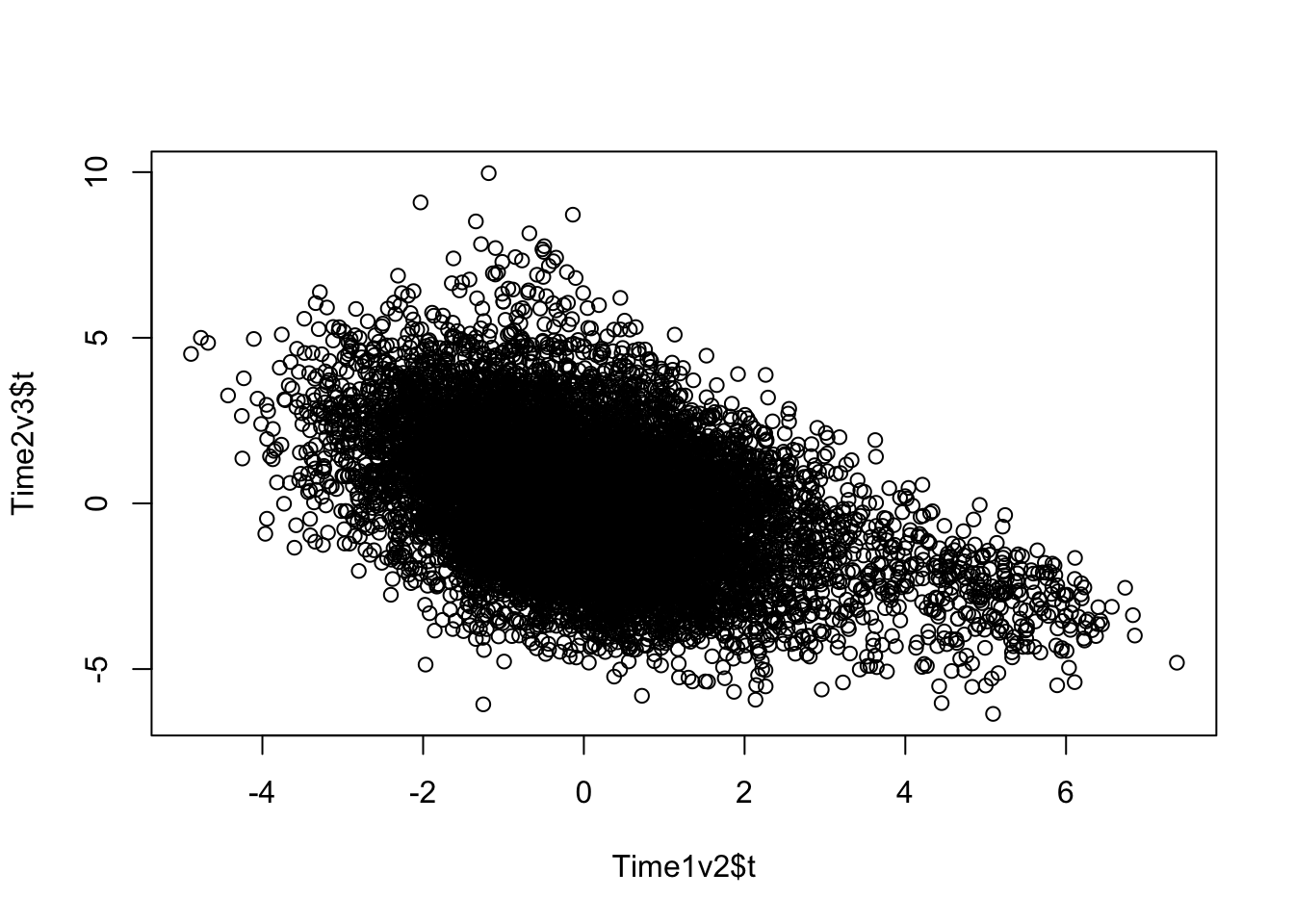
Expand here to see past versions of unnamed-chunk-2-11.png:
| Version | Author | Date |
|---|---|---|
| 7bcd123 | Lauren Blake | 2018-11-15 |
plot(Time1v2$adj.P.Val, Time2v3$adj.P.Val)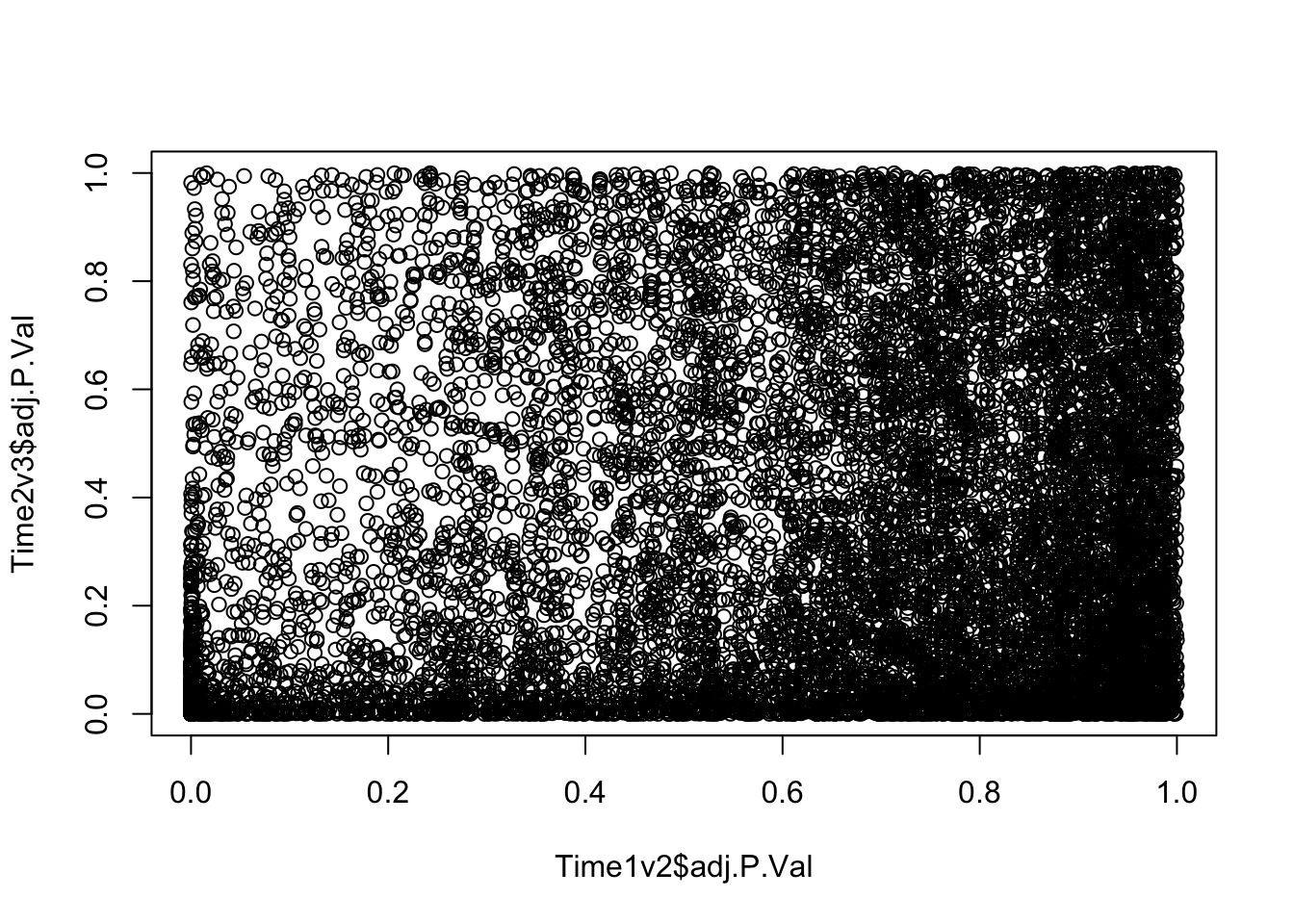
Expand here to see past versions of unnamed-chunk-2-12.png:
| Version | Author | Date |
|---|---|---|
| 7bcd123 | Lauren Blake | 2018-11-15 |
dim(Time1v2[which(Time1v2$adj.P.Val < FDR_level),])[1] 545 7dim(Time2v3[which(Time2v3$adj.P.Val < FDR_level),])[1] 2272 7head(topTable(fit1, coef=1, adjust="BH", number=100, sort.by="T")) genes logFC AveExpr t P.Value adj.P.Val
DCAF6 DCAF6 0.6665640 5.416600 7.379580 1.431162e-11 1.645551e-07
RNF10 RNF10 1.0377194 8.797228 6.855830 2.272978e-10 9.869232e-07
CTNNAL1 CTNNAL1 1.4096235 1.933041 6.831796 2.575030e-10 9.869232e-07
ALAS2 ALAS2 1.9585386 10.170073 6.735151 4.244316e-10 1.220029e-06
GPR146 GPR146 1.4288917 4.655669 6.569697 9.908589e-10 2.278579e-06
VTI1B VTI1B 0.5255125 6.179033 6.447501 1.841373e-09 3.086557e-06
B
DCAF6 15.90792
RNF10 13.24952
CTNNAL1 12.13729
ALAS2 12.60619
GPR146 11.85371
VTI1B 11.30170head(topTable(fit1, coef=2, adjust="BH", number=100, sort.by="T")) genes logFC AveExpr t P.Value
MPHOSPH8 MPHOSPH8 0.7255845 6.149600 9.968821 6.678677e-18
DFFA DFFA 0.5295244 4.523835 9.085363 1.091156e-15
RTF1 RTF1 0.5746842 5.542158 8.715696 8.936463e-15
PURA PURA 0.7886715 2.727101 8.512411 2.813599e-14
RP11-83A24.2 RP11-83A24.2 1.0124542 2.878809 8.153240 2.094359e-13
ZNF791 ZNF791 0.5775827 4.143783 7.827481 1.262969e-12
adj.P.Val B
MPHOSPH8 7.679143e-14 29.98284
DFFA 6.273054e-12 24.98292
RTF1 3.425048e-11 23.04615
PURA 8.087690e-11 21.35605
RP11-83A24.2 4.816188e-10 19.59222
ZNF791 2.420270e-09 18.21632Make a Venn Diagram of the DE genes
# FDR 1%
FDR_level <- 0.01
Time12 <- rownames(Time1v2[which(Time1v2$adj.P.Val < FDR_level),])
Time23 <- rownames(Time2v3[which(Time2v3$adj.P.Val < FDR_level),])
mylist <- list()
mylist[["DE T1 to T2"]] <- Time12
mylist[["DE T2 to T3"]] <- Time23
# Make as pdf
Four_comp <- venn.diagram(mylist, filename= NULL, main="DE genes between timepoints (1% FDR)", cex=1.5 , fill = NULL, lty=1, height=2000, width=2000, rotation.degree = 180, scaled = FALSE, cat.pos = c(0,0))
grid.draw(Four_comp)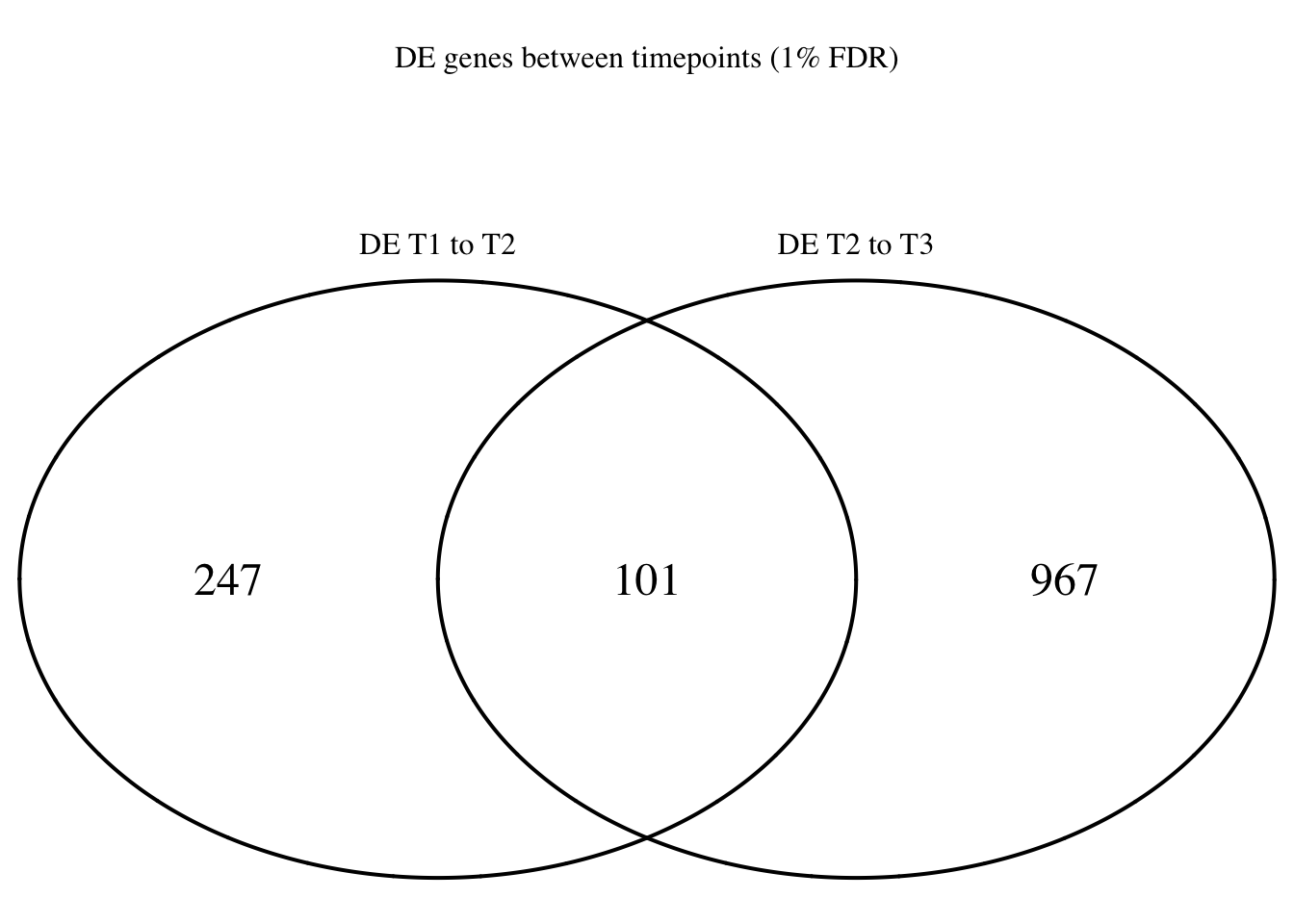
Expand here to see past versions of unnamed-chunk-3-1.png:
| Version | Author | Date |
|---|---|---|
| 7bcd123 | Lauren Blake | 2018-11-15 |
dev.off()null device
1 #pdf(file = "~/Dropbox/Figures/DET1_T2_change_weight_FDR1.pdf")
# grid.draw(Four_comp)
#dev.off()# FDR 5%
FDR_level <- 0.05
Time12 <- rownames(Time1v2[which(Time1v2$adj.P.Val < FDR_level),])
Time23 <- rownames(Time2v3[which(Time2v3$adj.P.Val < FDR_level),])
mylist <- list()
mylist[["DE T1 to T2"]] <- Time12
mylist[["DE T2 to T3"]] <- Time23
# Make as pdf
Four_comp <- venn.diagram(mylist, filename= NULL, main="DE genes between timepoints (5% FDR)", cex=1.5 , fill = NULL, lty=1, height=2000, width=2000, rotation.degree = 180, scaled = FALSE, cat.pos = c(0,0))
grid.draw(Four_comp)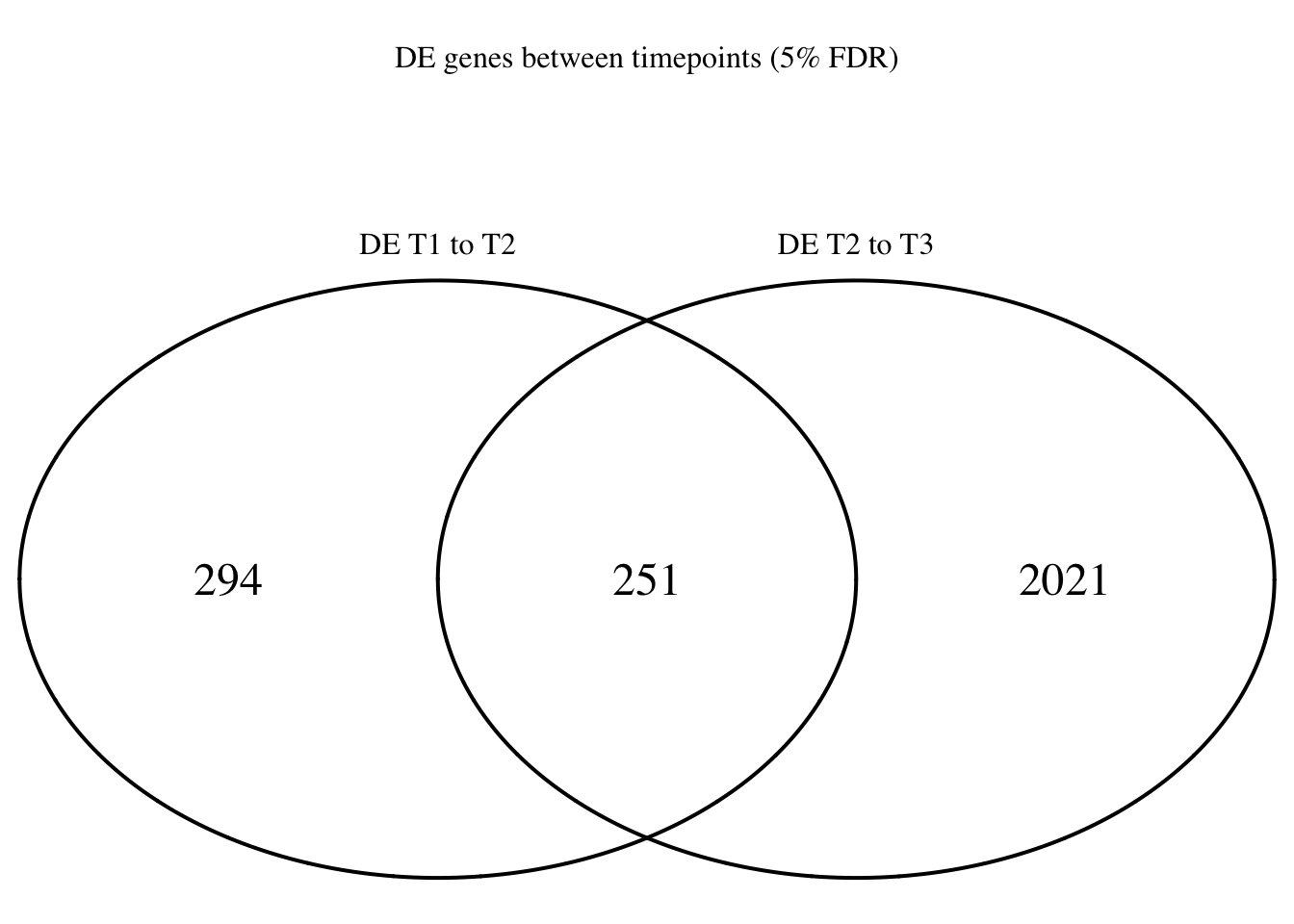
Expand here to see past versions of unnamed-chunk-4-1.png:
| Version | Author | Date |
|---|---|---|
| 7bcd123 | Lauren Blake | 2018-11-15 |
dev.off()null device
1 #pdf(file = "~/Dropbox/Figures/DET1_T2_change_weight_FDR5.pdf")
# grid.draw(Four_comp)
#dev.off()Session information
sessionInfo()R version 3.4.3 (2017-11-30)
Platform: x86_64-apple-darwin15.6.0 (64-bit)
Running under: OS X El Capitan 10.11.6
Matrix products: default
BLAS: /Library/Frameworks/R.framework/Versions/3.4/Resources/lib/libRblas.0.dylib
LAPACK: /Library/Frameworks/R.framework/Versions/3.4/Resources/lib/libRlapack.dylib
locale:
[1] en_US.UTF-8/en_US.UTF-8/en_US.UTF-8/C/en_US.UTF-8/en_US.UTF-8
attached base packages:
[1] grid stats graphics grDevices utils datasets methods
[8] base
other attached packages:
[1] cowplot_0.9.3 ggplot2_3.0.0 VennDiagram_1.6.20
[4] futile.logger_1.4.3 edgeR_3.20.9 limma_3.34.9
loaded via a namespace (and not attached):
[1] Rcpp_0.12.18 bindr_0.1.1 compiler_3.4.3
[4] pillar_1.3.0 formatR_1.5 git2r_0.23.0
[7] plyr_1.8.4 workflowr_1.1.1 R.methodsS3_1.7.1
[10] futile.options_1.0.1 R.utils_2.7.0 tools_3.4.3
[13] digest_0.6.16 evaluate_0.11 tibble_1.4.2
[16] gtable_0.2.0 lattice_0.20-35 pkgconfig_2.0.2
[19] rlang_0.2.2 yaml_2.2.0 bindrcpp_0.2.2
[22] withr_2.1.2 stringr_1.3.1 dplyr_0.7.6
[25] knitr_1.20 tidyselect_0.2.4 locfit_1.5-9.1
[28] rprojroot_1.3-2 glue_1.3.0 R6_2.2.2
[31] rmarkdown_1.10 purrr_0.2.5 lambda.r_1.2.3
[34] magrittr_1.5 whisker_0.3-2 backports_1.1.2
[37] scales_1.0.0 htmltools_0.3.6 assertthat_0.2.0
[40] colorspace_1.3-2 labeling_0.3 stringi_1.2.4
[43] lazyeval_0.2.1 munsell_0.5.0 crayon_1.3.4
[46] R.oo_1.22.0
This reproducible R Markdown analysis was created with workflowr 1.1.1